Abstract
Many plants, including Arabidopsis, increase in freezing tolerance in response to low, nonfreezing temperatures, a phenomenon known as cold acclimation. Previous studies established that cold acclimation involves rapid expression of the CBF transcriptional activators (also known as DREB1 proteins) in response to low temperature followed by induction of the CBF regulon (CBF-targeted genes), which contributes to an increase in freezing tolerance. Here, we present the results of transcriptome-profiling experiments indicating the existence of multiple low-temperature regulatory pathways in addition to the CBF cold response pathway. The transcript levels of ∼8000 genes were determined at multiple times after plants were transferred from warm to cold temperature and in warm-grown plants that constitutively expressed CBF1, CBF2, or CBF3. A total of 306 genes were identified as being cold responsive, with transcripts for 218 genes increasing and those for 88 genes decreasing threefold or more at one or more time points during the 7-day experiment. These results indicate that extensive downregulation of gene expression occurs during cold acclimation. Of the cold-responsive genes, 48 encode known or putative transcription factors. Two of these, RAP2.1 and RAP2.6, were activated by CBF expression and thus presumably control subregulons of the CBF regulon. Transcriptome comparisons indicated that only 12% of the cold-responsive genes are certain members of the CBF regulon. Moreover, at least 28% of the cold-responsive genes were not regulated by the CBF transcription factors, including 15 encoding known or putative transcription factors, indicating that these cold-responsive genes are members of different low-temperature regulons. Significantly, CBF expression at warm temperatures repressed the expression of eight genes that also were downregulated by low temperature, indicating that in addition to gene induction, gene repression is likely to play an integral role in cold acclimation.
INTRODUCTION
Many plants increase in freezing tolerance in response to low, nonfreezing temperatures, a phenomenon known as cold acclimation. Interest in understanding the molecular basis of cold acclimation is driven by both a desire to understand the mechanisms that plants have evolved to tolerate environmental stresses and the prospect that such knowledge might provide new strategies to improve the environmental stress tolerance of agriculturally important plants. Toward these ends, a number of important insights have been gained.
One is that the membrane systems of the cell are a primary site of freeze-induced injury and that this injury results largely from the severe dehydration associated with freezing (Levitt, 1980; Steponkus, 1984). Cellular dehydration can directly cause the formation of deleterious nonbilayer membrane structures such as hexagonal II–phase lipids. In addition, it can cause the generation of reactive oxygen species that can damage membranes and other cellular components. Therefore, cold acclimation must include the activation of mechanisms that protect cells against the potentially deleterious consequences of dehydration.
Multiple mechanisms appear to be involved, including changes in lipid composition and the accumulation of compatible solutes with cryoprotective properties, such as Suc and Pro (Thomashow, 1999). In addition, a number of cold-responsive genes encode hydrophilic LEA or LEA-like polypeptides thought to have roles in dehydration tolerance. Many studies provide support for this notion (Thomashow, 1999), including results indicating that the Arabidopsis COR (cold-regulated) gene COR15a encodes a novel hydrophilic polypeptide that acts directly as a cryoprotective protein that helps prevent the formation of hexagonal II–phase lipids (Steponkus et al., 1998).
A recent important insight into the cold acclimation process was the discovery of the Arabidopsis CBF cold response pathway (Shinozaki and Yamaguchi-Shinozaki, 2000; Thomashow, 2001). The promoters of many cold- and dehydration-responsive genes in Arabidopsis have been shown to contain a DNA regulatory element, the CRT (C-repeat)/DRE (dehydration-responsive element) (Baker et al., 1994; Yamaguchi-Shinozaki and Shinozaki, 1994), which confers both cold- and dehydration-responsive gene expression (Yamaguchi-Shinozaki and Shinozaki, 1994). A family of AP2-domain transcriptional activators, known as either the CBF (CRT binding factor) (Stockinger et al., 1997; Gilmour et al., 1998) or DREB1 (DRE binding) proteins (Liu et al., 1998; Shinwari et al., 1998), bind to the CRT/DRE element and activate transcription.
Three members of the CBF/DREB1 family, CBF1, CBF2, and CBF3 (or DREB1b, DREB1c, and DREB1a, respectively), are induced within 15 min of transferring plants to cold temperatures, followed at ∼2 h by expression of the CBF regulon of target genes, which are those genes whose promoters contain the CRT/DRE regulatory element (Gilmour et al., 1998; Liu et al., 1998; Shinwari et al., 1998). The CBF regulon includes genes that act in concert to improve freezing tolerance. Overexpression of the CBF/DREB1 transcription factors in transgenic Arabidopsis plants results in the accumulation of compatible solutes that have cryoprotective activities, including Pro, Suc, and raffinose (Gilmour et al., 2000). In addition, the CBF regulon includes the COR15a gene and others that encode LEA or LEA-like hydrophilic polypeptides thought to play roles in freezing tolerance.
Overexpression of the CBF/DREB1 proteins in Arabidopsis results in an increase in freezing tolerance at the whole-plant level in both nonacclimated and cold-acclimated plants (Jaglo-Ottosen et al., 1998; Liu et al., 1998; Kasuga et al., 1999; Gilmour et al., 2000) and enhances the tolerance of plants to dehydration caused by either imposed water deficit or exposure to high salinity (Liu et al., 1998; Kasuga et al., 1999). Recent studies (Jaglo et al., 2001) indicate that the CBF cold-response pathway is conserved in Brassica napus and that components of the pathway are present in wheat and rye, which cold acclimate, as well as in tomato, which does not.
The CBF cold response pathway is one low-temperature gene network that contributes to cold tolerance in Arabidopsis. A fundamental question is whether other low-temperature gene networks contribute significantly to freezing tolerance or other aspects of growth and development at low temperature. To address this issue, we used Affymetrix GeneChip arrays to analyze the Arabidopsis transcriptome at multiple times after transfer of plants from warm to cold temperature and in warm-grown plants that constitutively express CBF1, CBF2, or CBF3.
The results indicate that a dynamic series of changes in the Arabidopsis transcriptome is set in motion upon transfer of plants from warm to cold temperature, including changes in cold-regulatory gene networks in addition to the CBF cold response pathway. The results identify >250 newly described cold-responsive genes that offer explanations for certain biochemical changes that occur during cold acclimation, identify candidate polypeptides with roles in cold tolerance, and indicate that gene repression may play an integral role in the cold acclimation response.
RESULTS
Transfer of Arabidopsis to Low Temperature Produces Waves of Changes in the Transcriptome
Affymetrix GeneChip arrays, which contain 8297 DNA oligonucleotide probe sets representing ∼8000 genes per chip, were used to assay changes in the Arabidopsis transcriptome in response to exposure of plants to low temperature. Transcript levels were analyzed in duplicate biological samples harvested just before plants were transferred from 22 to 4°C (warm sample) and then at 0.5, 1, 4, 8, and 24 h and 7 days after transfer. Fold change values were calculated for the duplicate samples harvested at 4°C compared with each of the two warm samples, generating four comparisons for each time point.
A gene was designated as being upregulated at a given time point if the signal intensity was greater than background (“present”) for both duplicate cold samples, if there was a difference call of “increase” for all four comparisons, and if the fold increase value was ≥3 for all four comparisons. Similarly, a gene was designated as being downregulated at a given time point if the transcript levels produced a hybridization intensity greater than background for both duplicate warm samples, if there was a difference call of “decrease” for all four comparisons, and if the fold decrease value was ≥3 for all four comparisons.
Control experiments indicated that using a cutoff of threefold would effectively exclude the possibility of a gene being inappropriately designated as cold responsive as a result of a technical error. When a single RNA sample from warm-grown plants was used to prepare two probes that were hybridized to two different GeneChips, only 3 of the 8297 total probe sets were found to have a difference call of increase or decrease and a fold change value of ≥3, corresponding to a false-positive rate of 0.04%. Thus, using four comparisons to select cold-responsive genes, the false-positive rate would be predicted to be <2 per 1014 genes.
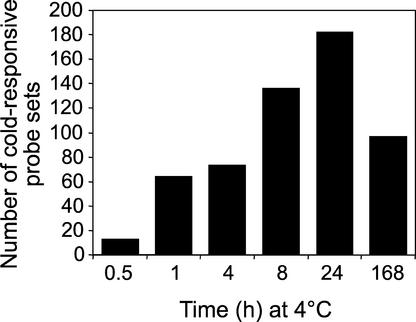
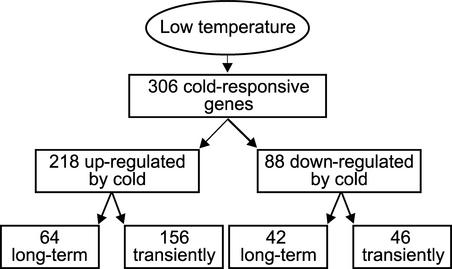
Using these criteria, 330 probe sets were found to represent cold-responsive genes, corresponding to 4% of the total probe sets. The number of cold-responsive probe sets increased to a maximum of 182 at 24 h and then decreased to 97 at 7 days (Figure 1). Analysis of these data indicated that 306 genes (some genes are represented by more than one probe set) were cold responsive at a minimum of one time point during the course of the experiment (Figure 2 and supplemental data online). Of these, 218 genes were scored as being upregulated in response to low temperature, and another 88 were scored as being downregulated (Figure 2).
Figure 1.
Number of GeneChip Probe Sets Representing Genes That Were Either Upregulated or Downregulated at Various Times after Transfer of Plants from Warm (22°C) to Cold (4°C) Temperature.
Figure 2.
Summary of Classes of Cold-Responsive Genes.
Details of selection criteria are described in the text. The total number of upregulated genes listed (218) is less than 156 (transient) + 64 (long-term) because probe sets representing two genes were present in both the transient and long term lists.
As expected, among the cold-regulated genes were members of the CBF cold response pathway. In particular, the transcript levels for CBF1, CBF2, and CBF3 increased within the first hour of plants being exposed to low temperature, followed closely (within 4 h) by expression of known CBF target genes, including COR6.6, ERD10, COR47, and COR78 (Figure 3). These results indicated that the tissue samples used in this experiment underwent a typical cold acclimation response and that the GeneChips replicated results obtained previously by RNA gel blot analysis (Gilmour et al., 1998; Liu et al., 1998; Shinwari et al., 1998).
Figure 3.
GeneChip Results for Genes Reported Previously as Upregulated during Cold Acclimation.
Probe sets used to calculate mean average difference values were as follows: COR47, probe sets 13225_s_at and 15997_s_at; ERD10, probe set 15103_s_at; COR78, probe set 15611_s_at; and COR6.6, probe sets 18699_i_at, 18700_r_at, and 18701_s_at. Where multiple probe sets were present that corresponded to a single gene, the mean average difference obtained for all corresponding probe sets was plotted.
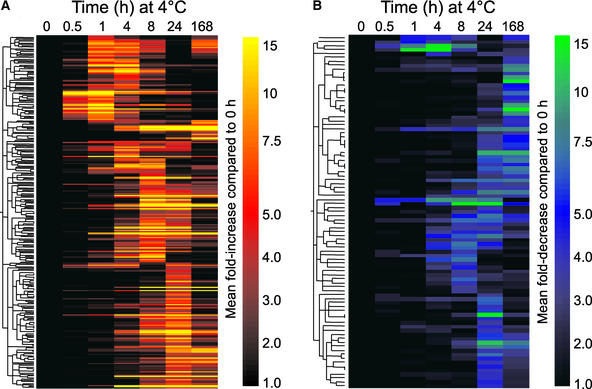
Hierarchical clustering of the entire set of 218 upregulated genes revealed that blocks of genes were induced in multiple waves after transfer of plants to low temperature (Figure 4A). In addition, it was evident that some genes were expressed transiently, whereas others were induced and remained activated for the entire 7-day experiment. Overall, the kinetic pattern observed did not fit a simple two-step cascade profile of the CBF cold response pathway—rapid cold induction of the CBF transcriptional activators (during the first hour) followed by expression of the CBF regulon (by 4 h)—suggesting that multiple regulatory pathways were activated in response to the temperature decrease. A similar picture emerged with the set of 88 downregulated genes (Figure 4B).
Figure 4.
Hierarchical Clustering of Cold-Responsive Genes.
The fold change values for genes that were upregulated (A) (n = 241 probe sets representing 218 genes) or downregulated (B) (n = 89 probe sets representing 88 genes) during cold acclimation (see Methods) were preprocessed so that fold change values that were associated with a difference call of no change were converted to 1. The mean of the four fold change values for each time point then was calculated, and the data were clustered using a Pearson correlation. Scales indicating the color assigned to each fold change are shown to the right of each cluster.
Long-Term Upregulated Genes Encode Multiple Transcription Factors and Proteins with Potential Roles in Freezing Tolerance
Transfer of plants from warm to cold temperature triggers the cold acclimation response, which includes expression of COR and other genes that remain upregulated for extended periods at low temperature. Of the 218 genes that were determined to be upregulated in response to cold (i.e., that were upregulated at least 3-fold at one or more time points during the course of the experiment), 64 genes remained upregulated at least 2.5-fold at 7 days. These were considered long-term upregulated genes (Figures 5A and 6 and supplemental data online). The lower threshold of 2.5-fold was used for this specific designation because we considered it inappropriate to assert that an upregulated gene showing expression at this level at 7 days had returned to a prestress level of expression. A search of the literature revealed that 50 of these long-term upregulated genes had not been reported previously to be cold regulated in Arabidopsis (supplemental data online).
Figure 5.
Hierarchical Clustering of Genes Upregulated by Cold.
Fold change values that were associated with a difference call of no change were converted to 1. The mean fold change values for each time point then were calculated, and the data were clustered. A scale indicating the color assigned to each fold change is shown to the right of the cluster.
(A) Clustering of the 64 genes (represented by 72 probe sets) that were upregulated by at least 2.5-fold after 7 days of cold treatment.
(B) Clustering of the 156 genes (represented by 169 probe sets) that were upregulated 3-fold at any time between 30 min and 24 h but were upregulated by less than 2.5-fold after 7 days of cold treatment.
Figure 6.
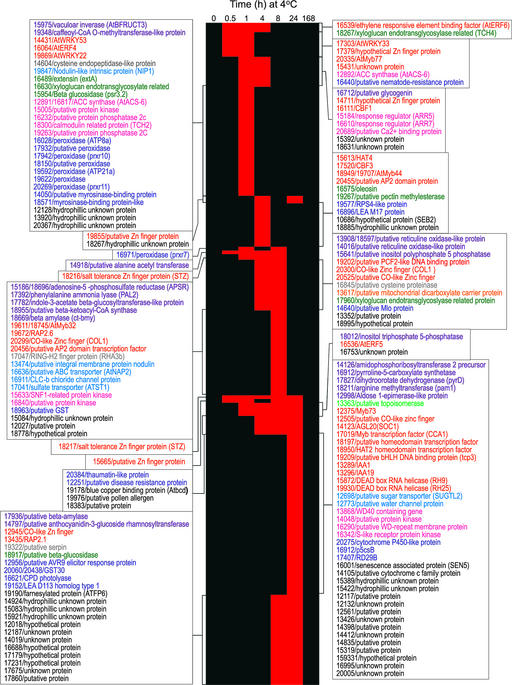
“Binary” Hierarchical Clustering of Long-Term Upregulated Genes.
Data points at which the signal intensity indicated that the gene was present for both duplicate cold samples, that there was a difference call of increase for all four comparisons, and that the fold increase value was ≥2.5 for all four comparisons were assigned a value of 2 (red), whereas all other data points were assigned a value of 1 (black). The resulting data then were clustered. The probe set number and the description of the genes that fall into each cluster are indicated at right. The text color indicates the known or predicted role of each gene: metabolism, purple; cell growth, cell division, and DNA synthesis, light green; transcription, red; protein fate, gray; transport facilitation, light blue; intracellular transport, orange; cellular biogenesis, dark green; cellular communication and signal transduction, pink; cell rescue, defense, cell death, and aging, dark blue; unclassified proteins, black.
Hierarchical clustering indicated that the 64 long-term upregulated genes were induced at different times after transfer of plants to low temperature (Figures 5A and 6). This was visualized most easily using a “binary” hierarchical clustering format (Figure 6). In this case, genes that were upregulated 2.5-fold or more were considered to be induced, and time points at which this occurred were colored red, whereas those that were not changed by 2.5-fold were considered unchanged, and these time points were colored black (thus, all time 0 values were black). The presentation reveals that blocks of genes were induced at each time point after transfer of plants to low temperature, suggesting that multiple regulatory pathways likely were involved in their induction.
Consistent with this notion was the finding that in addition to CBF2, eight other long-term cold-responsive genes that encoded either known or putative transcription factors were induced at various times during the course of the experiment (Figure 6 and supplemental data online). Two of these genes were induced rapidly in response to low temperature in parallel with CBF2, RAV1 (Kagaya et al., 1999) and ZAT12 (Meissner and Michael, 1997), which encode, respectively, an AP2 DNA binding factor and a zinc finger protein, the functions of which are not known. RNA gel blot analysis confirmed that transcripts for RAV1 and ZAT12 accumulated within 1 h of the transfer of plants to low temperature (Figure 7A).
Six additional long-term upregulated genes were found to encode known or putative transcription factors: a second putative zinc finger protein (At4g38960; Mayer et al., 1999), the R2R3-Myb transcription factor AtMYB73 (Kranz et al., 1998), H-protein promoter binding factor 2a (H.K.R. Abbaraju and D.J. Oliver, direct submission, AF079503), the HD-Zip protein AthB-12 (Lee and Chun, 1998), and two AP2 domain proteins, RAP2.7 and RAP2.1 (Okamuro et al., 1997) (Figure 6 and supplemental data online). Hierarchical clustering indicated that the expression patterns of these genes fell into four different groups (Figure 6), suggesting that they were regulated by multiple pathways.
The fact that these six genes were induced after the initial wave of CBF2, RAV1, and ZAT12 induction raised the possibility that one or more of them might be induced by one of these transcription factors. Inspection of the promoter region of RAP2.1 indicated that it contained two copies of the CCGAC core sequence of the CRT/DRE elements, suggesting that it might be a target of the CBF activators. Indeed, RNA gel blot analysis indicated that the transcript levels of RAP2.1 did not increase until 4 to 8 h after transfer of plants to low temperature (Figure 7A) and that they were increased in transgenic Arabidopsis plants that constitutively overexpressed CBF1, CBF2, or CBF3 (Figure 7B). Thus, the CBF regulon presumably includes a “subregulon” controlled by RAP2.1.
In addition to long-term upregulated genes encoding transcription factors, 55 genes encoded proteins with diverse known or proposed functions (Figure 6 and supplemental data online). The largest group encoded COR/LEA proteins, polypeptides thought to play roles in cryoprotection (Thomashow, 1999). In addition to the previously described COR6.6, COR15b, COR47, COR78, and ERD10 polypeptides was the dehydrin Xero2, a LEA3-type protein designated Di21, and a polypeptide (At1g01470) with a high degree of sequence similarity to group 4 LEA proteins (Terryn et al., 1998).
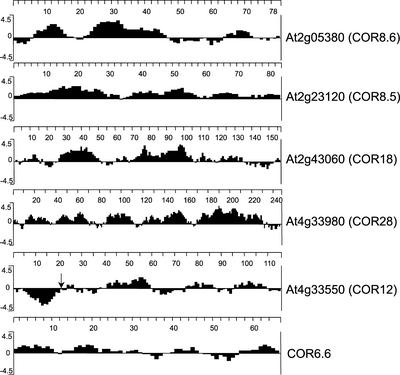
All of these COR/LEA proteins have different sequences, but they have in common the biochemical property of being highly hydrophilic. Interestingly, 5 of the 11 long-term upregulated unknown proteins also encoded polypeptides that were highly hydrophilic (Figure 8). We designated these COR/LEA-like polypeptides COR8.5 (At2g23120), COR8.6 (At2g05380), COR12 (At4g33550), COR18 (At2g43060), and COR28 (At4g33980), which encode polypeptides of 8.5, 8.6, 12.3, 17.8, and 28.1 kD, respectively. The N-terminal end of COR12 is predicted to encode a signal sequence that would result in the secretion of a mature hydrophilic polypeptide of 10.1 kD.
Figure 8.
Hydropathy Plots for Novel COR-Like Proteins.
The amino acid sequence predicted from the sequence of COR-like proteins was analyzed using the method of Kyte and Doolittle (1982) to predict the regional hydropathy of the encoded polypeptides. Values > 0 correspond to hydrophilic regions, and values < 0 correspond to hydrophobic regions. The scale at top of each plot shows the number of amino acids from the N terminus of the polypeptide. The polypeptide encoded by At4g33550 is predicted to have a signal peptide (iPSORT; www.HypothesisCreator.net/iPSORT/) that is cleaved where indicated by the arrow. The hydropathy profile of COR6.6 is shown for comparison.
Sugars, including Suc and raffinose, accumulate during cold acclimation in Arabidopsis (Wanner and Junttila, 1999; Gilmour et al., 2000). Thus, genes that encode proteins with roles in sugar metabolism might be expected to be cold responsive. We found previously that transcripts encoding Suc synthase accumulated in response to low temperature (Gilmour et al., 2000), a finding that is confirmed here (Figure 6 and supplemental data online). Additionally, transcripts for three different genes encoding putative galactinol synthases (At1g09350, At2g47180, and At1g60470), the enzyme that catalyzes the first committed step in the synthesis of raffinose, were found to accumulate in response to low temperature. In one case, induction was >160-fold at 24 h (Figure 6 and supplemental data online). Transcripts encoding a putative sugar transporter also accumulated in response to low temperature.
Among the most rapid and highly induced genes was that encoding ELIP1 (Early Light-Induced Protein 1) (Figure 6 and supplemental data online). ELIP1, as well as ELIP2, are nuclear-encoded thylakoid membrane proteins that are expressed in response to light stress (Moscovici-Kadouri and Chamovitz, 1997; Heddad and Adamska, 2000). They are thought to be photoprotective pigment carriers or chlorophyll exchange proteins (Adamska, 1997). Expression of this gene indicates that the plants potentially experienced light stress.
Finally, an intriguing finding was that transcripts for GIGANTEA (GI) were found to increase 5- to 10-fold in response to low temperature (Figure 6 and supplemental data online). The GI gene encodes a protein that has no homology with any proteins of known function in the databases (Fowler et al., 1999; Park et al., 1999). Although the function of the GI protein is unknown, mutations in the GI gene cause a pleiotropic phenotype with effects on flowering in response to photoperiod, phytochrome B signaling, the circadian clock, and carbohydrate metabolism (Koornneef et al., 1991; Eimert et al., 1995; Fowler et al., 1999; Park et al., 1999; Huq et al., 2000), but no association with cold acclimation has been reported.
Transient Upregulated Genes Encode Proteins with a Wide Range of Functions and Suggest a Period of Oxidative Stress after Transfer of Plants to Low Temperature
Of the 306 genes designated cold responsive, 156 (51%) were upregulated transiently in response to low temperature (Figures 5B and 9 and supplemental data online). Hierarchical clustering of these genes revealed that transfer of plants from warm to cold temperature set off a series of transient waves of changes in the transcriptome (Figures 5B and 9). At each time point, new genes were upregulated, and in most cases, they remained induced for only one or two of the time points. As with the long-term cold-responsive genes, this pattern was more complex than the two-step CBF cold response pathway, suggesting that multiple regulatory systems were involved in the response to temperature decrease.
Figure 9.
“Binary” Hierarchical Clustering of Transiently Upregulated Genes.
Data points at which the signal intensity indicated that the gene was present for both duplicate cold samples, that there was a difference call of increase for all four comparisons, and that the fold increase value was ≥3 for all four comparisons were assigned a value of 2 (red), whereas all other data points were assigned a value of 1 (black). The resulting data then were clustered. The probe set number and the description of the genes that fall into each cluster are indicated at right. The text color indicates the known or predicted role of each gene: metabolism, purple; cell growth, cell division, and DNA synthesis, light green; transcription, red; protein fate, gray; transport facilitation, light blue; intracellular transport, orange; cellular biogenesis, dark green; cellular communication and signal transduction, pink; cell rescue, defense, cell death, and aging, dark blue; unclassified proteins, black.
Indeed, of the 156 transiently cold-induced genes, 34 (22%) corresponded to known or putative transcription factors (Figure 9 and supplemental data online). In addition, 16 genes (10%) encoded known or putative proteins involved in signal transduction or cellular communication, including response regulators, protein kinases, and phosphatases. In total, ∼33% of the transiently expressed genes potentially had roles in gene regulation.
The transient nature of the changes in transcript levels suggested that the abrupt temperature decrease might have resulted in a short-lived “shock” response followed by an adjustment to the new environmental conditions. Indeed, low temperature can cause a decrease in the turnover rate of photosystem II components, causing an increase in photosystem II excitation pressure, or “excess excitation energy,” and the generation of damaging reactive oxygen species, including hydrogen peroxide (Huner et al., 1998).
An indication that such a response occurred in our experiments was the fact that among the transiently expressed genes were three known or putative glutathione S-transferases, which are known to be involved in the detoxification of toxic metabolites arising from oxidative damage caused by excess excitation energy (Marrs, 1996), and nine known or putative peroxidases that potentially also contribute to the detoxification of hydrogen peroxide (Østergaard et al., 1998). Moreover, seven genes that have been shown to be induced by hydrogen peroxide in Arabidopsis (Desikan et al., 2001) were among the genes we found to be induced transiently by cold. These genes encoded a blue copper binding protein, adenosine-5-phosphosulfate reductase, a putative zinc finger transcription factor (At5g04340), AtERF-4, CCA1, a putative nematode resistance protein, and a protein of unknown function (At2g36220).
The production of ethylene also has been associated with cold stress (Ciardi et al., 1997; Morgan and Drew, 1997; Yu et al., 2001). In this regard, it was notable that genes involved in ethylene signaling were among those that were induced rapidly with low temperature (Figure 9 and supplemental data online). Within the first hour of transfer, transcripts accumulated for 1-aminocyclopropane-1-carboxylic acid synthase (AtACS-6), which catalyzes a limiting step in ethylene synthesis, and two ethylene-responsive transcription factors, AtERF-4 and AtERF-5 (Fujimoto et al., 2000).
However, two other known ethylene-inducible genes, AtERF-1 and basic chitinase, were not induced (these genes are represented by probe sets on the GeneChip). This and the fact that cold induction of AtERF-4 and AtERF-5 can occur independent of ethylene (Fujimoto et al., 2000) led us to conclude that although rapid transfer of plants to low temperature resulted in a burst of transcript accumulation for 1-aminocyclopropane-1-carboxylic acid synthase, little if any ethylene actually was produced.
Of the total transiently expressed genes, 24% encoded unknown proteins (Figure 9 and supplemental data online). Of these, 11 were novel polypeptides that, like LEA/COR proteins, were unusually hydrophilic. The probe sets corresponding to these genes were At2g41010, At1g23710, At2g28400, At4g35320, At2g22860, At4g04330, At1g10410, At2g46970, At2g32210, At2g36220, and At1g11960 (supplemental data online). In addition, transcript levels for genes encoding three previously described LEA/COR proteins (LEA M17, LEA D113, and RD29B) were found to be expressed transiently (Figure 9 and supplemental data online). These results indicate that low temperature generated a short-lived signal that induces the expression of LEA/COR and LEA/COR-like genes through a pathway that is independent of the CBF cold response pathway.
Genes Encoding Proteins with Diverse Functions Are Downregulated during Cold Acclimation
Of the 306 genes that were scored as being cold responsive, 88 (27%) corresponded to genes that were downregulated, 46 of which were affected transiently and 42 of which were affected long term (i.e., they were downregulated 2.5-fold or more at 7 days) (Figure 2 and supplemental data online). This is a clear indication that extensive downregulation of gene expression occurs in response to low temperature.
Like the genes that were upregulated in response to low temperature, the downregulated genes encoded proteins with a wide range of functions, including transcription, signaling, cell wall biogenesis, and defense. Four of the long-term downregulated transcripts encoded proteins with known or predicted roles in energy production: two light-harvesting proteins, Lhca2*1 and Lhcb4*3; a putative 5-kD photosystem II protein; and a ferredoxin precursor. The downregulation of these genes may have resulted from the decreased light levels that the plants were exposed to during the cold treatment. However, it has been reported that the transfer of warm-grown plants to chilling temperature (4°C) leads to a rapid inhibition of photosynthesis followed by a reduction in transcript levels for genes encoding photosynthetic proteins (Krapp and Stitt, 1995; Strand et al., 1997). This effect of low temperature may have caused the decrease in transcript levels of photosynthesis-related proteins observed here.
A striking difference between the downregulated and upregulated genes was that few genes were downregulated during the first 4 h of plant exposure to cold temperature. The full significance of this apparent delay is uncertain, because any decrease in transcript levels must take into account the turnover rate of the transcript. Given that the half-life of the average plant transcript is on the order of several hours (Abler and Green, 1996), one would not expect many changes until after 4 h. However, most (68%) of the changes did not occur until the 24-h time point or after, indicating that even when the turnover rate of transcripts is taken into account, the mechanisms that lead to the downregulation of transcript levels in response to cold treatment are delayed compared with those that lead to upregulation.
Genes Regulated by the CBF Cold Response Pathway Are a Subset of Total Cold-Responsive Genes
Many of the findings described above indicate that regulatory pathways in addition to the CBF cold response pathway are activated in response to low temperature. To explore this issue further, we profiled the transcriptomes of warm-grown transgenic Arabidopsis plants that constitutively expressed CBF1, CBF2, or CBF3 and compared them with the profiles of control plants. Fold change values were calculated by comparing the data for each transgenic line against those for two control samples, generating six comparisons.
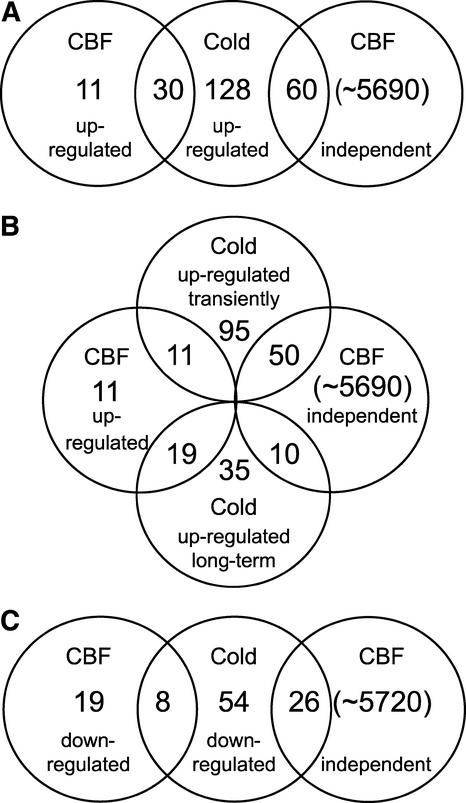
A gene was designated as being a member of the CBF regulon if the signal intensity was greater than background (present) for all three transgenic lines, if there was a difference call of increase for all six comparisons, and if the fold increase value was ≥3 for all six comparisons. For a gene to be designated as cold responsive but independent of the CBF cold response pathway, it had to be upregulated by cold treatment but could not be upregulated in any of the CBF-overexpressing plants; that is, the probe sets had to be assigned a difference call of “no change” in each of the six comparisons between the three CBF transgenic lines and two individual wild-type samples. These criteria were stringent; consequently, 128 (59%) of the genes upregulated by cold could not be assigned to either of the categories. However, for 90 genes, an assignment was made.
Of the cold-induced genes, 60 were not upregulated in any of the CBF-expressing plants; thus, they were designated as being independent of the CBF cold response pathway (Figure 10A). Of these, the large majority, 50 genes (or 83%), were upregulated transiently upon exposure to low temperature; the remaining 10 (17%) were long-term upregulated (Figure 10B).
Figure 10.
Venn Diagrams of Comparisons between Cold-Responsive Genes and Genes That Are Part of the CBF Regulon.
Sets of genes were selected using the criteria described in Methods. The number of genes in each set is displayed within a circle above a description of the set. Genes present in two sets are shown in the intersection of the two sets, so that the sum of the numbers within a circle is the total number of genes in that set.
(A) Intersection of genes that are upregulated in response to low temperature with those that are either upregulated by or independent of CBF overexpression.
(B) Intersection of genes that are either transiently or long-term upregulated in response to low temperature with those that are either upregulated by or independent of CBF overexpression.
(C) Intersection of genes that are downregulated in response to low temperature with those that are either downregulated by or independent of CBF overexpression.
Analysis of the CBF-expressing plants indicated that 41 genes were upregulated in all three transgenic lines (Figure 10A and supplemental data online). Included in this group were genes reported previously as being CBF targets, such as COR6.6, COR78, COR47, P5CSb, and ERD10 (Jaglo-Ottosen et al., 1998; Gilmour et al., 2000; Seki et al., 2001). Of the 41 CBF-regulated genes, 30 were found to be upregulated in response to low temperature in the experiments described above (Figure 10A and supplemental data online). Thus, these 30 genes were considered genuine members of the CBF regulon. The 11 CBF-responsive genes that were not cold induced presumably were downstream consequences of CBF expression related to the altered growth phenotypes displayed by plants that overexpressed CBF (Liu et al., 1998; Kasuga et al., 1999; Gilmour et al., 2000). Of the 30 genes that were members of the CBF regulon, 19 were in the long-term cold-responsive group and the remaining 11 were induced transiently (Figure 10B). Among these genes was RAP2.6, which encodes an AP2 protein and thus is another likely transcription factor that controls the expression of a subregulon of the CBF regulon.
Finally, of the 88 genes found to be downregulated during cold acclimation, 8 also were downregulated by CBF overexpression: At4g22690, At3g57260, At1g75040, At2g14560, At1g21270, At2g43570, At1g69490, and At4g14400 (Figures 10C and supplemental data online). These results indicate that the CBF cold-response pathway not only acts to activate gene expression but also is involved in repressing the expression of certain genes.
DISCUSSION
Central goals in cold acclimation research include identifying cold-responsive genes, determining how they are regulated, and understanding their roles in plant life at low temperature. Most of the studies to date have been with individual or small numbers of genes. However, with the development of genomic technologies, including methods for gene expression profiling, these issues can be addressed on a more global scale.
Seki et al. (2001) recently used such methods to analyze the expression of 1300 Arabidopsis genes using a cDNA microarray. Their experiments resulted in the identification of 19 cold-inducible genes, 10 of which were newly described. Nine of the 19 cold-induced genes were shown to be part of the CBF regulon (i.e., they were induced in response to DREB1a expression). Moreover, 15 of the cold-inducible genes also were induced in response to drought.
This report expands on the findings of Seki et al. (2001) by describing the expression of ∼8000 genes at multiple times after transfer of plants from warm to low temperature. The results provide an unprecedented view of the flux that occurs in the Arabidopsis transcriptome upon the shifting of plants from warm to cold temperature. Within 30 min of the transfer of plants to low temperature, waves of changes in the composition of the transcriptome are initiated, and these continue to develop beyond 24 h. A total of 306 genes were found to be affected, the large majority of which (to the best of our knowledge) have not been described previously as being cold responsive in Arabidopsis (Figures 2, 6, and 9 and supplemental data online).
Both increases and decreases in transcript levels occurred, some of which were transient and others of which were long lived, being sustained for up to 7 days of cold treatment. The affected genes encompassed a wide range of functions, including transcription, signaling, metabolism, cellular biogenesis, and cell rescue and defense. The results indicate that the expression of as much as 4% of the genome may be affected by exposing plants to low temperature. Thus, if the probe sets used in our experiments generally are representative of the entire genome, then ∼1000 genes would be predicted to be cold responsive. This probably is a low estimate, however, because we used stringent criteria to designate a gene as being cold responsive. We considered only those transcript levels that had increased or decreased at least threefold in each of the four comparisons of the duplicate samples. In addition, genes that are expressed at low levels may have been excluded. Indeed, ∼20% of the probe sets gave no signal at any time during the experiment.
Fifteen of the 19 genes that Seki et al. (2001) identified as being cold inducible were represented by probe sets on the Affymetrix GeneChips used in our experiments. Six of these genes (COR47, COR78, COR6.6, ERD10, SEN12/ERD7, and CBF3/DREB1a) had been reported previously to be cold inducible and were identified as cold regulated in our experiments. One of the nine novel cold-regulated genes identified by Seki et al. (2001), which encodes a β-amylase (FL5-90/At4g17090), also was found by us to be cold regulated.
Four additional novel cold-regulated genes identified by Seki et al. (2001) were upregulated by cold in our experiments (were given a difference call of increase) but were not above our threefold cutoff and thus were not designated as being cold-inducible. These genes, which were upregulated between twofold and threefold in the experiments reported by Seki et al. (2001), encoded a putative cold acclimation protein (FL3-5A3/At2g15970), a nodulin-like protein (FL5-1A9/At4g27520), ferritin (FL5-3A15/At5g01600), and a homolog of DC1.2 (FL5-2I22/At5g62350).
Finally, four of the genes designated as being cold regulated by Seki et al. (2001) were not cold regulated in our experiments. These genes, which were induced between twofold and threefold, encoded a homolog of the LEA protein SAG21 (FL5-3M24/At4g02380), a rice glyoxalase homolog (FL5-95/At1g11840), a DEAD box ATPase/RNA helicase (FL2-5A4/At3g01540), and EXGT-A2 (FL5-3P12/At1g14720). The reason for this difference is not known but could reflect differences in plant culture conditions, environmental treatments, or the expression-profiling methods used.
Previous studies have established that the CBF cold response pathway is an integral component of the cold acclimation response (Shinozaki and Yamaguchi-Shinozaki, 2000; Thomashow, 2001). Additional cold-regulatory pathways also might have important roles in cold tolerance, contributing to increased freezing tolerance and mediating the physiological, biochemical, and structural changes required for growth and development at low temperature. Indeed, Xin and Browse (1998) described a mutant of Arabidopsis, designated eskimo1, which is constitutively freezing tolerant but does not express known members of the CBF regulon.
Our results provide direct evidence for cold-regulatory pathways in addition to the CBF cold response pathway. Of the 306 cold-responsive genes, 106 were affected over the long term (i.e., transcript levels were upregulated or downregulated at least 2.5-fold at 7 days). Among these genes were members of the CBF cold response pathway. However, in addition, eight other genes encoding known or putative transcription factors were found to be long-term cold responsive: ZAT12, RAV1, AtMYB73, ATHB-12, H-protein binding factor 2a, RAP2.1, a zinc finger protein (At4g38960), and RAP2.7 (Figure 6 and supplemental data online).
One of these, RAP2.1, proved to be a target of CBF transcription factors (Figure 7B) and presumably regulates the expression of a subregulon of genes within the larger CBF regulon. However, two of the transcription factors, ZAT12 and RAV1, were found to be induced in parallel with the CBF transcriptional activators (i.e., transcript levels were increased more than threefold at 30 min). It is possible that the low-temperature regulation of ZAT12 and RAV1 involves the action of the same regulatory proteins that activate CBF expression. Gilmour et al. (1998) previously speculated on the existence of such a “super regulon” controlled by the hypothetical protein ICE (Inducer of CBF Expression).
Figure 7.
Transcript Levels for Cold-Regulated Transcription Factors RAV1, ZAT12, and RAP2.1.
(A) Two-week-old wild-type (Wassilewskija-2) plants grown at 22°C were cold treated at 4°C, and tissue was harvested at the times indicated. Total RNA was isolated, and RNA gel blots were prepared (10 μg of RNA). The blots were hybridized with 32P-labeled probes for RAV1, ZAT12, and RAP2.1.
(B) Total RNA was isolated from 2-week-old plants from transgenic lines expressing the indicated CBF genes under the control of the 35S promoter of Cauliflower mosaic virus or carrying the empty vector (V). Total RNA was isolated from plants grown at warm temperature, and RNA gel blots were prepared (10 μg) and hybridized with a 32P-labeled probe for RAP2.1.
Regardless, the parallel induction of CBF2, ZAT12, and RAV1, well before the induction of known CBF target genes such as COR47, COR6.6, and COR78, argues against their being members of the CBF regulon. Consistent with this notion was the fact that ZAT12 transcript levels were unaffected in CBF1-, CBF2-, or CBF3-overexpressing plants. A comparison of the transcriptomes of CBF-overexpressing plants and control plants indicated that at least 60 cold-induced genes are independent of the CBF cold response pathway, including at least 15 transcription factors.
Investigations to date have focused on genes that are upregulated in response to low temperature. The results presented here, however, indicate that downregulation of gene expression also may be an important component of the adaptation to low temperature. Indeed, the downregulation response was extensive. Transcript levels for 88 genes were found to decrease either transiently or long term in response to low temperature. These genes, like those that were upregulated, encompassed a wide range of functions, including transcription, signaling, cell wall biogenesis, defense, and photosynthesis, almost all of which (to the best of our knowledge) have not been shown previously to be cold repressed.
Some of the changes in transcript levels, especially those associated with photosynthesis, might have been caused by the lower light conditions used during the cold treatment. However, this would not appear to be true for all of the down-regulated genes. In particular, eight genes that were downregulated in response to low temperature also were downregulated at warm temperature in response to the constitutive expression of CBF1, CBF2, and CBF3. These data indicate that the CBF cold response pathway includes downregulation of the expression of certain genes and suggest genes whose expression might be incompatible with the enhancement of freezing tolerance.
Previous studies have identified 14 genes as members of the CBF regulon. Here, we expand that number to 45 (37 upregulated and 8 downregulated). Among the newly described members of the CBF regulon are genes encoding two putative transcription factors, RAP2.1 and RAP2.6, that presumably control the expression of subregulons of the larger CBF regulon. In this regard, it is interesting that a number of the genes that are activated by CBF expression do not have the core CCGAC sequence of the CRT/DRE element within 1 kb of the start of transcription. These are candidate members of a CBF subregulon controlled by RAP2.1, RAP2.6, or a yet-to-be-discovered transcription factor(s) induced by the CBF activators.
Other new members of the CBF regulon include genes that encode a putative sugar transporter, water channel proteins, and a new hydrophilic polypeptide that potentially acts as a cryoprotectant. In addition, there are three genes that putatively encode galactinol synthase, which catalyzes the first committed step in raffinose synthesis. In fact, while we were preparing this article, Taji et al. (2002) reported that three Arabidopsis genes encode proteins with galactinol synthase activity and that one of these, AtGolS3 (which corresponds to probe set 18596_at), was induced in response to low temperature and overexpression of CBF3/DREB1a.
Given that we sampled approximately one-third of the genome in our experiments, there could be as many as 100 genes that are part of the CBF regulon. Again, this is probably an underestimate because the criteria used here to assign a gene to the CBF regulon included it being induced threefold or more in each CBF overexpression line. However, it is not known whether CBF1, CBF2, and CBF3 control completely overlapping sets of genes. We observed differences in the set of genes that were upregulated in the CBF1, CBF2, and CBF3 transgenic plants tested here. However, we do not know whether these differences were attributable to biological variation, differences in the level of CBF expression, or bona fide differences in the activities of the CBF proteins. Additional experiments will be required to address this issue definitively.
One intriguing finding is that among the 280 newly described cold-responsive genes was GI. GI transcript levels increased fivefold to eightfold after 24 h of cold treatment and remained increased after 7 days. GI is a novel protein with roles in the promotion of flowering by photoperiod and circadian clock function (Fowler et al., 1999; Park et al., 1999), but it has not been associated with acclimation to low temperature. Interestingly, however, it is known that in addition to entrainment by light/dark cycles, the circadian clock in plants can be entrained by temperature cycles (Kloppstech et al., 1991; Beator et al., 1992; Heintzen et al., 1994; McWatters et al., 2000). Park et al. (1999) proposed that GI functions in a light input pathway to the clock. The observation that GI is cold responsive raises the possibility that GI might function in input pathways to the clock that transmit both light and temperature signals.
More than half of the cold-responsive genes were upregulated or downregulated transiently in response to low temperature. It seems likely that the expression of a significant number of these genes was caused by the abrupt change in temperature used in our experiments. We transferred plants directly from 22 to 4°C, a protocol that is used commonly in the study of cold acclimation. Transferring plants from warm to cold temperature in the light (as was done in our experiments) can result in excess light energy, which can lead to the production of hydrogen peroxide and other reactive oxygen species (Huner et al., 1998). This, in turn, can lead to the induction of genes involved in the protection of cells against oxidative stress.
A number of genes known to be responsive to hydrogen peroxide were upregulated transiently in our experiments, including glutathione S-transferases, indicating that the plants at least transiently experienced oxidative stress. Thus, some of the transient cold-responsive genes probably do not play direct roles in life at low temperature per se but instead play critical roles in enabling plants to adjust to quickly fluctuating environmental conditions, including protection against conditions that result in excess light (Huner et al., 1998). It would not be appropriate, however, to draw the conclusion that none of the transiently expressed genes plays a role in freezing tolerance or other fundamental aspects of life at low temperature. CBF1 and CBF3 were among the transiently expressed genes; thus, inclusion on this list certainly cannot be used to dismiss a gene's potential importance in cold acclimation. It also is true that arbitrary criteria were used to place genes into the transient category; their transcript levels had to have returned to within 2.5-fold that of the warm sample after 7 days of cold treatment. However, the transcript levels for ∼55% of these genes were increased 2.5-fold or more at 24 h, so it is possible that many of them remained actively upregulated for several days after transfer to the cold. Moreover, the levels of the proteins for these genes could have remained increased for many days after transfer to cold.
Finally, it is relevant that when Escherichia coli (and other bacteria) are subjected to an abrupt temperature decrease of ∼15°C, they undergo a cold-shock response (Yamanaka, 1999) that includes the transient induction of genes, some of which (e.g., the Csp gene family) have been shown to play critical roles in enabling bacteria to grow at low temperature (Xia et al., 2001).
The results of this transcriptome study demonstrate the highly complex nature of plant adaptation to low temperature. The results indicate that the expression of hundreds of genes is affected upon exposure of plants to low temperature and that this involves the action of multiple cold-regulatory pathways, including the activation of regulons within regulons. Through the further application of genomic approaches, it should be possible to construct a diagram of the low-temperature “gene circuitry” in plants and to determine the roles of the regulatory networks in cold tolerance.
METHODS
Plant Material and Growth Conditions
Arabidopsis thaliana ecotype Wassilewskija-2 and transgenic plants constitutively expressing CBF1 (G5, G6, and G26), CBF2 (E2, E8, and E24; S.J. Gilmour and M.F. Thomashow, unpublished results), or CBF3 (A28, A30, and A40; Gilmour et al., 2000) in the Wassilewskija-2 background were used in these experiments. Seeds were surface-sterilized and then spread on Petri plates containing Gamborg's B-5 medium (Life Technologies, Gaithersburg, MD) solidified with 0.8% phytagar (Life Technologies). Immediately after plating, the seeds were stratified for 4 days at 4°C to ensure uniform germination.
Plants were grown in controlled-environment chambers at 22°C under continuous illumination of 100 μmol·m−2·s−1 from cool-white fluorescent lights for 11 days. For cold treatments, plates containing the plants were transferred to 4°C under continuous light (20 to 40 μmol·m−2·s−1) as described (Gilmour et al., 1998), and tissue samples were harvested after 0.5, 1, 4, 8, and 24 h and 7 days. Duplicate samples for each cold time point and single samples from each of the CBF transgenic lines were harvested for profiling.
Upon request, all novel material described in this article will be made available in a timely manner for noncommercial research purposes. No restrictions or conditions will be placed on the use of any materials described in this article that would limit their use for noncommercial research purposes.
RNA Isolation and Probe Labeling
The aerial parts of 50 to 150 plants grown on a single plate were pooled for each RNA sample. Total RNA was extracted from the samples using the RNeasy Plant Kit (Qiagen, Valencia, CA). Biotinylated target RNA was prepared from 16 μg of total RNA using the procedure described by the manufacturer of the Arabidopsis GeneChip (Affymetrix, Santa Clara, CA). Briefly, a primer encoding a T7 RNA polymerase promoter fused to (dT)24 (Genset Oligos, La Jolla, CA) was used to prime double-stranded cDNA synthesis using the SuperScript Choice System (Life Technologies). The resulting cDNA was transcribed in vitro using the BioArray High-Yield RNA Transcript Labeling Kit (Enzo Biochem, New York, NY) in the presence of biotinylated UTP and CTP to produce biotinylated target complementary RNA (cRNA).
Affymetrix GeneChip Hybridization and Data Collection
The labeled target cRNA was purified, fragmented, and hybridized to Arabidopsis Genome GeneChip arrays according to protocols provided by the manufacturer (Affymetrix) in a Hybridization Oven model 640 (Affymetrix). The GeneChips were washed and stained with streptavidin-phycoerythrin using a GeneChip Fluidics Station model 400 and then scanned with a Gene Array Scanner (Hewlett-Packard, Palo Alto, CA).
Data Analysis
The Microarray Suite 4.0 and Data Mining Tool 1.0 (Affymetrix) software packages were used for the analysis of microarray data. The output from all GeneChip hybridizations was scaled globally such that its average intensity was equal to an arbitrary target intensity of 100. Because all experiments were scaled to the same target intensity, comparison between GeneChips was possible. The mean noise for the GeneChips used in these analyses was 3.7 ± 3.5. Average difference (gene expression) and fold change values were calculated from the GeneChip fluorescent intensity data.
The software also was used to determine whether expression of each gene was present or absent (absolute call) and whether the fold change value represented a genuine change in expression (difference call). Fold change values were calculated for each sample harvested at 4°C compared with each of the samples harvested before transfer to 4°C, generating four measurements for each gene at each time point during the cold treatment. Fold change values also were calculated for the three CBF transgenic samples compared with each of the wild-type samples, generating six measurements for each gene.
Probe sets that met the following criteria were selected for further analysis. Those determined to be upregulated by the cold treatment were selected as having, at any time point, an absolute call of present in both cold samples, difference calls of increase, and fold changes of at least 3.0 for all four fold change comparisons. Those determined to be downregulated by the cold treatment were selected as having, at any time point, an absolute call of present in both warm samples, difference calls of decrease, and fold changes of at least −3.0 for all four fold change comparisons.
Cold-regulated genes were determined to be long-term upregulated if, at the 7-day time point, both absolute calls were present and the four fold change comparisons were 2.5-fold or greater associated with four difference calls of increase. Similarly, cold-regulated genes were determined to be long-term downregulated if, at the 7-day time point, the fourfold change comparisons were −2.5-fold or greater associated with four difference calls of decrease.
Genes upregulated by CBF overexpression were selected as having an absolute call of present in all three CBF transgenic samples, difference calls of increase, and a fold change of at least 3.0 for all six fold change comparisons between wild-type and CBF transgenic lines. Those determined to be downregulated by CBF expression were selected as having an absolute call of present in both wild-type samples, difference calls of decrease, and fold changes of at least −3.0 for all six fold change comparisons between wild-type and CBF transgenic lines. Cold-regulated genes that were independent of CBF expression were selected as having a difference call of no change in all six fold change comparisons between wild-type and CBF transgenic lines.
Microsoft Access database management software (Microsoft, Redmond, WA) was used to manage and filter the GeneChip data, and Genespring 4.0.4 (Silicon Genetics, Redwood City, CA) was used to generate hierarchical gene clusters using a Pearson correlation (separation ratio of 0.5, minimum distance of 0.001). Before clustering, all data points that were associated with a difference call of no change were converted to 1. Binary hierarchical clusters were generated by altering the data used to generate the clusters such that data points that fulfilled a particular set of criteria were converted to 2, whereas all other points were assigned a value of 1.
The false-positive rate was calculated as the number of probe sets changed significantly as a percentage of probe sets on the array (Lipshutz et al., 1999). A single total RNA sample was used to prepare two cDNA samples and subsequently cRNA samples, which then were hybridized to two different GeneChips and fold change values were calculated. Genes were counted as false changes if they showed changes of threefold or greater associated with a difference call of increase and a signal threshold greater than background (present) in at least one sample of the comparison.
RNA Gel Blot Hybridization Analysis
Total RNA samples (10 μg), isolated as described above, were electrophoresed on agarose gels, and RNA gel blot transfers were prepared and hybridized as described in Hajela et al. (1990) using high-stringency wash conditions (Stockinger et al., 1997). Gene-specific probes to RAV1, RAP2.1, and ZAT12 were obtained by amplifying ∼1 kb of the coding region of these genes from Arabidopsis genomic DNA The primers used were as follows: RAV1, 5′-TCTAGACGA-AAAAGTCGTCGGTAGGT-3′ and 5′-GGATCCGAGTTGTTACGA-GGCGTGAA-3′; ZAT12, 5′-ACTAGTCAGAAGAAAAATGGTTGC-GATA-3′ and 5′-GGATCCGAAAAATTCAAAGAATGAGAGAAACA-3′; and RAP2.1, 5′-TCTAGATCAATGGAAAGAGAACAAGAA-3′ and 5′-AGATCTAAATTGACTATATATCTCCGGATTC-3′. The probes were radiolabeled with 32P by priming with random octamers (Invitrogen, Carlsbad, CA).
Supplementary Material
Acknowledgments
We thank Dan Zarka for preparing the RNA samples used in the GeneChip experiments; Dan Zarka and Jonathon Vogel for performing the RNA gel blot analysis shown in Figure 7; Steve Triezenberg and Sarah Gilmour for critical comments on the manuscript; Michael Gieseg, Li Tian, and Annette Thelen for helpful discussions and advice; and Marlene Cameron for help with figures. GeneChip hybridization and data collection were performed at the Genomics Technology Support Facility (Michigan State University). This work was supported by grants from the National Science Foundation (Grant DBI 0110124), the Department of Energy (Grant DEFG0291ER20021), and the Michigan Agricultural Experiment Station.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.003483.
Footnotes
Online version contains Web-only data.
References
- Abler, M.L., and Green, P.L. (1996). Control of mRNA stability in higher plants. Plant Mol. Biol. 32, 63–78. [DOI] [PubMed] [Google Scholar]
- Adamska, I. (1997). ELIPs: Light induced stress proteins. Physiol. Plant. 100, 794–805. [Google Scholar]
- Baker, S.S., Wilhelm, K.S., and Thomashow, M.F. (1994). The 5′-region of Arabidopsis thaliana cor15a has cis-acting elements that confer cold-, drought- and ABA-regulated gene expression. Plant Mol. Biol. 24, 701–713. [DOI] [PubMed] [Google Scholar]
- Beator, J., Pötter, E., and Kloppstech, K. (1992). Coordinated circadian regulation of mRNA levels for light-regulated genes and of the capacity for accumulation of chlorophyll protein complexes. Plant Physiol. 100, 1780–1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciardi, J.A., Deikman, J., and Orzolek, M.D. (1997). Increased ethylene synthesis enhances chilling tolerance in tomato. Physiol. Plant. 101, 333–340. [Google Scholar]
- Desikan, R., A.-H.-Mackerness, S., Hancock, J.T., and Neill, S.J. (2001). Regulation of the Arabidopsis transcriptome by oxidative stress. Plant Physiol. 127, 159–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eimert, K., Wang, S.-M., Lue, W.-L., and Chen, J. (1995). Monogenic recessive mutations causing both late floral initiation and excess starch accumulation in Arabidopsis. Plant Cell 7, 1703–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler, S., Lee, K., Onouchi, H., Samach, A., Richardson, K., Morris, B., Coupland, G., and Putterill, J. (1999). GIGANTEA: A circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J. 18, 4679–4688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto, S.Y., Ohta, M., Usui, A., Shinshi, H., and Ohme-Takagi, M. (2000). Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box–mediated gene expression. Plant Cell 12, 393–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour, S.J., Sebolt, A.M., Salazar, M.P., Everard, J.D., and Thomashow, M.F. (2000). Overexpression of the Arabidopsis CBF3 transcriptional activator mimics multiple biochemical changes associated with cold acclimation. Plant Physiol. 124, 1854–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour, S.J., Zarka, D.G., Stockinger, E.J., Salazar, M.P., Houghton, J.M., and Thomashow, M.F. (1998). Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. Plant J. 16, 433–442. [DOI] [PubMed] [Google Scholar]
- Hajela, R.K., Horvath, D.P., Gilmour, S.J., and Thomashow, M.F. (1990). Molecular cloning and expression of cor (cold-regulated) genes in Arabidopsis thaliana. Plant Physiol. 93, 1246–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heddad, M., and Adamska, I. (2000). Light stress-regulated two-helix proteins in Arabidopsis thaliana related to the chlorophyll a/b-binding gene family. Proc. Natl. Acad. Sci. USA 97, 3741–3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzen, C., Melzer, S., Fischer, R., Kappeler, S., Apel, K., and Staiger, D. (1994). A light- and temperature-entrained circadian clock controls expression of transcripts encoding nuclear proteins with homology to RNA-binding proteins in meristematic tissue. Plant J. 5, 799–813. [DOI] [PubMed] [Google Scholar]
- Huner, P.A., Öquist, G., and Sarhan, F. (1998). Energy balance and acclimation to light and cold. Trends Plant Sci. 3, 224–230. [Google Scholar]
- Huq, E., Tepperman, J.M., and Quail, P.H. (2000). GIGANTEA is a nuclear protein involved in phytochrome signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 97, 9789–9794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaglo, K.R., Kleff, S., Amundsen, K.L., Zhang, X., Haake, V., Zhang, J.Z., Deits, T., and Thomashow, M.F. (2001). Components of the Arabidopsis C-repeat/dehydration-responsive element binding factor cold-response pathway are conserved in Brassica napus and other plant species. Plant Physiol. 127, 910–917. [PMC free article] [PubMed] [Google Scholar]
- Jaglo-Ottosen, K.R., Gilmour, S.J., Zarka, D.G., Schabenberger, O., and Thomashow, M.F. (1998). Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 280, 104–106. [DOI] [PubMed] [Google Scholar]
- Kagaya, Y., Ohmiya, K., and Hattori, T. (1999). RAV1, a novel DNA-binding protein, binds to bipartite recognition sequence through two distinct DNA-binding domains uniquely found in higher plants. Nucleic Acids Res. 27, 470–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasuga, M., Liu, Q., Miura, S., Yamaguchi-Shinozaki, K., and Shinozaki, K. (1999). Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat. Biotechnol. 17, 287–291. [DOI] [PubMed] [Google Scholar]
- Kloppstech, K., Otto, B., and Sierralta, W. (1991). Cyclic temperature treatments of dark-grown pea seedlings induce a rise in specific transcript levels of light-regulated genes related to photomorphogenesis. Mol. Gen. Genet. 225, 468–473. [DOI] [PubMed] [Google Scholar]
- Koornneef, M., Hanhart, C.J., and van der Veen, J.H. (1991). A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol. Gen. Genet. 229, 57–66. [DOI] [PubMed] [Google Scholar]
- Kranz, H.D., et al. (1998). Towards functional characterization of the members of the R2R3-MYB gene family from Arabidopsis thaliana. Plant J. 16, 263–276. [DOI] [PubMed] [Google Scholar]
- Krapp, A., and Stitt, M. (1995). An evaluation of direct and indirect mechanisms for the “sink-regulation” of photosynthesis in spinach: Changes in gas exchange, carbohydrates, metabolites, enzyme activities and steady-state transcript levels after cold-girdling source leaves. Planta 195, 313–323. [Google Scholar]
- Kyte, J., and Doolittle, R.F. (1982). A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157, 105–132. [DOI] [PubMed] [Google Scholar]
- Lee, Y.H., and Chun, J.Y. (1998). A new homeodomain-leucine zipper gene from Arabidopsis thaliana induced by water stress and abscisic acid treatment. Plant Mol. Biol. 37, 377–384. [DOI] [PubMed] [Google Scholar]
- Levitt, J. (1980). Responses of Plants to Environmental Stresses, 2nd ed. (New York: Academic Press).
- Lipshutz, R.J., Fodor, S.P., Gingeras, T.R., and Lockhart, D.J. (1999). High density synthetic oligonucleotide arrays. Nat. Genet. 21, 20–24. [DOI] [PubMed] [Google Scholar]
- Liu, Q., Kasuga, M., Sakuma, Y., Abe, H., Miura, S., Yamaguchi-Shinozaki, K., and Shinozaki, K. (1998). Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10, 1391–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrs, K.A. (1996). The functions and regulation of glutathione S-transferases in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47, 127–158. [DOI] [PubMed] [Google Scholar]
- Mayer, K., et al. (1999). Sequence and analysis of chromosome 4 of the plant Arabidopsis thaliana. Nature 402, 769–777. [DOI] [PubMed] [Google Scholar]
- McWatters, H.G., Bastow, R.M., Hall, A., and Millar, A.J. (2000). The ELF3 zeitnehmer regulates light signalling to the circadian clock. Nature 408, 716–720. [DOI] [PubMed] [Google Scholar]
- Meissner, R., and Michael, A.J. (1997). Isolation and characterization of a diverse family of Arabidopsis two and three-fingered C2H2 zinc finger protein genes and cDNAs. Plant Mol. Biol. 33, 615–624. [DOI] [PubMed] [Google Scholar]
- Morgan, P.W., and Drew, M.C. (1997). Ethylene and plant responses to stress. Physiol. Plant. 100, 620–630. [Google Scholar]
- Moscovici-Kadouri, S., and Chamovitz, D.A. (1997). Characterization of a cDNA encoding the early light-inducible protein (ELIP) (accession no. U89014) from Arabidopsis (PGR 97-155). Plant Physiol. 115, 1287.9390449 [Google Scholar]
- Okamuro, J.K., Caster, B., Villarroel, R., Van Montagu, M., and Jofuku, K.D. (1997). The AP2 domain of APETALA2 defines a large new family of DNA binding proteins in Arabidopsis. Proc. Natl. Acad. Sci. USA 94, 7076–7081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Østergaard, L., Pedersen, A.G., Jespersen, H.M., Brunak, S., and Welinder, K.G. (1998). Computational analyses and annotations of the Arabidopsis peroxidase gene family. FEBS Lett. 433, 98–102. [DOI] [PubMed] [Google Scholar]
- Park, D.H., Somers, D.E., Kim, Y.S., Choy, Y.H., Lim, H.K., Soh, M.S., Kim, H.J., Kay, S.A., and Nam, H.G. (1999). Control of circadian rhythms and photoperiodic flowering by the Arabidopsis GIGANTEA gene. Science 285, 1579–1582. [DOI] [PubMed] [Google Scholar]
- Seki, M., Narusaka, M., Abe, H., Kasuga, M., Yamaguchi-Shinozaki, K., Carninci, P., Hayashizaki, Y., and Shinozaki, K. (2001). Monitoring the expression pattern of 1300 Arabidopsis genes under drought and cold stresses by using a full-length cDNA microarray. Plant Cell 13, 61–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki, K., and Yamaguchi-Shinozaki, K. (2000). Molecular responses to dehydration and low temperature: Differences and cross-talk between two stress signaling pathways. Curr. Opin. Plant Biol. 3, 217–223. [PubMed] [Google Scholar]
- Shinwari, Z.K., Nakashima, K., Miura, S., Kasuga, M., Seki, M., Yamaguchi-Shinozaki, K., and Shinozaki, K. (1998). An Arabidopsis gene family encoding DRE/CRT binding proteins involved in low-temperature-responsive gene expression. Biochem. Biophys. Res. Commun. 250, 161–170. [DOI] [PubMed] [Google Scholar]
- Steponkus, P.L. (1984). Role of the plasma membrane in freezing injury and cold acclimation. Annu. Rev. Plant Physiol. 35, 543–584. [Google Scholar]
- Steponkus, P.L., Uemura, M., Joseph, R.A., Gilmour, S.J., and Thomashow, M.F. (1998). Mode of action of the COR15a gene on the freezing tolerance of Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 95, 14570–14575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockinger, E.J., Gilmour, S.J., and Thomashow, M.F. (1997). Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc. Natl. Acad. Sci. USA 94, 1035–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strand, Å., Hurry, V., Gustafsson, P., and Gardeström, P. (1997). Development of Arabidopsis thaliana leaves at low temperatures releases the suppression of photosynthesis and photosynthetic gene expression despite the accumulation of soluble carbohydrates. Plant J. 12, 605–614. [DOI] [PubMed] [Google Scholar]
- Taji, T., Ohsumi, C., Iuchi, S., Seki, M., Kasuga, M., Kobayashi, M., Yamaguchi-Shinozaki, K., and Shinozaki, K. (2002). Important roles of drought- and cold-inducible genes for galactinol synthase in stress tolerance in Arabidopsis thaliana. Plant J. 29, 417–426. [DOI] [PubMed] [Google Scholar]
- Terryn, N., Gielen, J., De Keyser, A., Van Den Daele, H., Ardiles, W., Neyt, P., De Clercq, R., Coppieters, J., Dehais, P., Villarroel, R., Rouze, P., and Van Montagu, M. (1998). Sequence analysis of a 40-kb Arabidopsis thaliana genomic region located at the top of chromosome 1. Gene 215, 11–17. [DOI] [PubMed] [Google Scholar]
- Thomashow, M.F. (1999). Plant cold acclimation, freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 571–599. [DOI] [PubMed] [Google Scholar]
- Thomashow, M.F. (2001). So what's new in the field of plant cold acclimation? Lots! Plant Physiol. 125, 89–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanner, L.A., and Junttila, O. (1999). Cold-induced freezing tolerance in Arabidopsis. Plant Physiol. 120, 391–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, B., Ke, H., and Inouye, M. (2001). Acquirement of cold sensitivity by quadruple deletion of the cspA family and its suppression by PNPase S1 domain in Escherichia coli. Mol. Microbiol. 40, 179–188. [DOI] [PubMed] [Google Scholar]
- Xin, Z., and Browse, J. (1998). eskimo1 mutants of Arabidopsis are constitutively freezing-tolerant. Proc. Natl. Acad. Sci. USA 95, 7799–7804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki, K., and Shinozaki, K. (1994). A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 6, 251–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka, K. (1999). Cold shock response in Escherichia coli. J. Mol. Microbiol. Biotechnol. 1, 193–202. [PubMed] [Google Scholar]
- Yu, X.M., Griffith, M., and Wiseman, S.B. (2001). Ethylene induces antifreeze activity in winter rye leaves. Plant Physiol. 126, 1232–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.