Abstract
Survival of Escherichia coli O157:H7 was investigated using a dynamic gastrointestinal model. A high bacterial mortality was observed in the stomach and duodenum. In contrast, bacteria grew in the distal parts of the small intestine. The coadministration of Saccharomyces cerevisiae CNCM I-3856 led to a significant reduction of bacterial resumption, maybe through ethanol production.
Enterohemorrhagic Escherichia coli (EHEC) is a food-borne pathogen that causes human diseases ranging from uncomplicated diarrhea to hemorrhagic colitis and hemolytic-uremic syndrome. Human contamination occurs mainly by ingestion of raw ground beef, vegetables, water, and dairy products contaminated by cattle feces (3, 25). Most of the outbreaks and sporadic cases of infection around the world are caused by EHEC strains that belong to serotype O157:H7 (24, 26). The virulence of EHEC strains is associated with the ability of the bacteria to attach to and efface intestinal brush borders and produce Shiga toxins Stx1 and/or Stx2. The terminal ileum and the colon are supposed to be the principal sites of EHEC colonization and pathology in humans (10, 15, 24).
To cause human illness, E. coli O157:H7 must survive the gastric and small intestinal environment in transit toward the large intestine. Gastric pH, gastrointestinal secretions, and peristalsis are well-known factors controlling the outcome of food-borne pathogens. However, few reports have really focused on the behavior of E. coli O157:H7 in the human gastrointestinal tract. The available data (1, 4, 27, 30) were obtained in gastric static systems that are not representative of the continuously changing variables during chyme transit. The TNO gastrointestinal tract model (TIM; Zeist, Netherlands) is an alternative dynamic multicompartmental in vitro system which presently allows the closest simulation of in vivo physiological processes occurring within the lumen of the stomach and small intestine of human (22). It consists of four successive compartments simulating the stomach, duodenum, jejunum, and ileum of human. The main parameters of digestion, such as pH, body temperature, peristaltic mixing and transport, gastric, biliary, and pancreatic secretions, and passive absorption of small molecules and water, are reproduced as accurately as possible. One of the main advantages of the TIM system is the possibility to collect digestive samples at any level of the gastrointestinal tract and at any time during digestion. This model has been validated for microbial applications and is particularly relevant for probiotics (6, 21) and pathogenic microorganism (18) survival studies.
Since antibiotic therapy is regarded as controversial in the treatment of EHEC infections (33), probiotics are being investigated as an alternative strategy. Probiotics are defined as live microorganisms that resist digestion and reach the colon alive and, when administered in adequate amounts, confer a health benefit on the host (14). To control EHEC infection, probiotic administration may be considered at the following two levels: (i) in ruminants to decrease E. coli O157:H7 carriage and therefore reduce the risk of human food-borne disease or (ii) in humans to exert a direct antagonist effect against the pathogen. Probiotic bacteria or yeasts have been evaluated for their capacity to reduce EHEC growth in batch cultures (2, 23), decrease the infectious proinflammatory response in infected T84 cells (12), or limit EHEC fecal shedding in cattle (32).
Until now, Saccharomyces cerevisiae var. boulardii was the only yeast commercialized as a probiotic for human use. S. cerevisiae CNCM I-3856 is a new marketed yeast probiotic (Lynside Pro GI+, Lesaffre Human Care, Milwaukee, WI) which has already shown anti-inflammatory activities in induced colitis in mice (16) and efficiency in reducing digestive discomfort and abdominal pain in patients with irritable bowel syndrome (IBS) (13).
In this work, we describe the use of the TIM system to evaluate the survival of an E. coli O157:H7 strain in simulated human gastrointestinal conditions and the influence of S. cerevisiae CNCM I-3856 on bacterial viability. To closely mimic physiological conditions, the pathogen (with or without yeast) was given in a typical Western diet containing raw ground beef, one of the main foods involved in E. coli O157:H7 outbreaks.
Artificial digestions in the TIM system.
An aerobic culture (LB, 24 h, 37°C) of an isogenic mutant of E. coli O157:H7 strain EDL 933 lacking the stx1 and stx2 genes (19) was used to inoculate the test meal (2.5 × 105 CFU/ml) prior to its introduction into the TIM system. This mutant has been widely used to study EHEC infectious processes (10, 20, 28). When coadministrated with bacteria, S. cerevisiae CNCM I-3856 was given in its active dried powder form (107 CFU/ml). The TIM system was programmed to reproduce the digestion of a solid meal in a healthy human adult (Table 1). The total duration of the digestions was 300 min (n = 4 digestions for each condition). Samples were taken in the test meal (initial intake) before its introduction into the artificial stomach and regularly collected during digestion in the different compartments of the system (stomach, duodenum, jejunum, and ileum). Microbial counting was performed on sorbitol-MacConkey agar (bacteria) and on Sabouraud dextrose agar (yeast). To evaluate bacterial and yeast survival rates in the TIM, control digestions (n = 4) were carried out in the same experimental conditions with water containing 0.8% (wt/vol) of a nonabsorbable transit marker, blue dextran (22). The concentrations of ethanol in the digestive samples were determined using an enzymatic UV test (Biosentec, Toulouse, France). Significant differences between treatments were tested by analysis of variance (ANOVA) with repeated-measure analysis followed by a post hoc test (SAS 9.1 Software, Inc., Cary, NC). P values of <0.05 were taken to indicate statistical differences.
TABLE 1.
Parameters of gastrointestinal digestion in the TIM system when simulating digestive conditions of a healthy adult after intake of a solid meala
| Compartment | Vol (ml) at initial time | pH/time (min) | Secretion | t1/2(min) | ß coefficient |
|---|---|---|---|---|---|
| Gastric | 300 | 2/0, 6/5, 5.7/15, 4.5/45, 2.9/90, 2.3/120, 1.8/240, 1.6/300 | 0.25 ml/min of pepsin (2,080 IU/ml), 0.25 ml/min of lipase (250.5 IU/ml), 0.25 ml/min of HCl (1.5 M) if necessary | 85 | 1.8 |
| Duodenal | 30 | Maintained at 6.0 | 0.5 ml/min of bile salts (4% during the first 30 min of digestion and then 2%), 0.25 ml/min of pancreatic juice (103 USP/ml), 0.25 ml/min of intestinal electrolyte solution, 0.25 ml/min of NaHCO3 (1 M) if necessary, 23,600 IU of trypsin (at the beginning of digestion) | ||
| Jejunal | 130 | Maintained at 6.9 | 0.25 ml/min of NaHCO3 (1 M) if necessary | ||
| Ileal | 130 | Maintained at 7.2 | 0.25 ml/min of NaHCO3 (1 M) if necessary | 250 | 2.5 |
The power exponential equation (f = 1 −  , where f represents the fraction of meal delivered, t the time of delivery, t1/2 the half-time of delivery, and β a coefficient describing the shape of the curve) is used for the computer control of gastric and ileal deliveries in the TIM.
, where f represents the fraction of meal delivered, t the time of delivery, t1/2 the half-time of delivery, and β a coefficient describing the shape of the curve) is used for the computer control of gastric and ileal deliveries in the TIM.
Survival of E. coli O157:H7 in simulated gastrointestinal conditions.
The viability of E. coli O157:H7 was evaluated in each TIM compartment by comparing the curves obtained for the bacteria and for the transit marker (Fig. 1). Our results showed that during its transit through the stomach (Fig. 1a) and duodenum (Fig. 1b) of the TIM, E. coli O157:H7 was largely killed, probably due to the occurrence of stringent conditions such as gastric acidity, digestive enzymes, and bile salts. In particular, bile salts are toxic at high concentrations for bacterial cells by disorganizing the lipid bi-layer structure of the cellular membranes (31). At the end of gastric digestion, 34% ± 4% of the initial bacteria are still alive and are in transit to the small intestine (Fig. 2). Even if acid resistance features have been described for E. coli O157:H7 strains (17), these bacteria were found to be sensitive to low pH in the present study. Indeed, cell mortality was observed in the artificial stomach from 60 min, when the pH fell below 4 (Fig. 2). Large variations in survival rates (from 0 to 100%) were observed for E. coli O157:H7 in in vitro gastric models (1, 4, 27, 29, 30). This wide range of values may be explained by differences between culture conditions, digestive systems (static or dynamic) and parameters, food matrices, and also bacterial strains. Interestingly, the only study that evaluated the behavior of E. coli O157:H7 in dynamic conditions (reproduction of the fall in gastric pH) gave survival percentages after gastric digestion close to those obtained in our study (29).
FIG. 1.
Influence of S. cerevisiae CNCM I-3856 on the survival of E. coli O157:H7 during in vitro digestions in the stomach (a), duodenum (b), jejunum (c), and ileum (d) of the TIM. The results obtained in each compartment for bacteria without (black bar) or with (white bar) yeast are compared with that of the transit marker, blue dextran (black line). Results are expressed as mean percentages ± standard deviations (SD) (n = 4 digestions) of initial intake. E. coli O157:H7 (*) and E. coli O157:H7 + S. cerevisiae (†) results significantly different from those of the transit marker (P < 0.05). E. coli O157:H7 results significantly different from those of E. coli O157:H7 plus S. cerevisiae at P values of <0.05 (+) or P values of <0.01 (++).
FIG. 2.
Cumulative gastric delivery of viable E. coli O157:H7 (thick black line) and blue dextran (thick gray line) in the TIM. Results are expressed as mean percentages ± SD (n = 4 digestions) of initial intake. The fall of gastric pH during in vitro digestions is plotted (thin black line). *, E. coli O157:H7 results significantly different from those of the transit marker (P < 0.05).
In the distal parts of the artificial gastrointestinal tract, growth resumption was observed at the end of digestion. At 300 min, the percentages for bacteria largely exceeded that of the transit marker in the jejunum (35% ± 14% versus 8% ± 1%) (Fig. 1c) and in the ileum (58% ± 19% versus 25% ± 1%) (Fig. 1d). This growth resumption was probably linked to less stringent environmental conditions, such as a pH closer to neutrality, lower concentrations of bile salts (owing to their passive reabsorption), and/or an increase in the residence time of bacteria. This study is the first report on the behavior of E. coli O157:H7 in human small-intestinal conditions. Bacterial growth renewal has been already observed in the small-intestinal compartments of the TIM but for a nonpathogenic strain of E. coli (18).
Antagonistic effect of S. cerevisiae CNCM I-3856.
The lack of specific treatment for EHEC infections has prompted work on alternative preventive and/or curative strategies, such as the use of probiotics. Among potential candidates, we evaluated the influence of a new probiotic S. cerevisiae strain on the viability of E. coli O157:H7 in the TIM system. The survival of E. coli O157:H7 in the stomach and duodenum of the artificial digestive system was not significantly modified by the coadministration of S. cerevisiae CNCM I-3856 (Fig. 1a and b). On the contrary, at the end of digestion (300 min), the addition of yeast led to a 10-fold decrease in bacterial survival in the jejunum (P = 0.0008) (Fig. 1c). Similarly, 58% ± 19% of the bacteria initially introduced in the TIM was recovered after 300 min of digestion in the ileum when bacteria were administered alone, whereas only 23% ± 7% of bacteria was found when yeasts were added in the test meal (P = 0.0057) (Fig. 1d). For the first time, we highlighted a direct antagonistic effect of S. cerevisiae against an EHEC strain in simulated human conditions. This interesting result suggests that this yeast could be used in humans to limit the amount of E. coli O157:H7 that reaches the distal small intestine and the colon. The antagonistic effect observed in vitro may be explained by (i) the competition between yeasts and bacteria for nutrient utilization, (ii) the modification by yeasts of the physico-chemical conditions of the digestive environment (i.e., redox potential), or (iii) the production by yeasts of inhibitory substances (11), such as proteases (7, 8) or ethanol.
To further investigate the antagonistic mechanisms between bacteria and yeast, ethanol concentrations were measured in the intestinal compartments of the TIM. While near-zero levels were observed in samples collected during digestions with E. coli O157:H7 alone, the ethanol contents reached 0.58 ± 0.15 g/liter when S. cerevisiae CNCM I-3856 was added in the test meal (P < 0.01) (Fig. 3). We therefore showed, for the first time, that an S. cerevisiae strain is able to produce ethanol directly in the human digestive environment. Ethanol, like other organic solvents, may be lethal for bacteria by disrupting the membrane to repress cell growth (9). Since yeasts synthesize ethanol from simple sugars released during digestion, it was not surprising to observe high levels of ethanol in the distal parts of the small intestine.
FIG. 3.
Ethanol concentrations in jejunal and ileal samples from the TIM. **, E. coli O157:H7 results significantly different from those of E. coli O157:H7 plus S. cerevisiae (P < 0.01).
Survival of S. cerevisiae CNCM I-3856 in simulated gastrointestinal conditions.
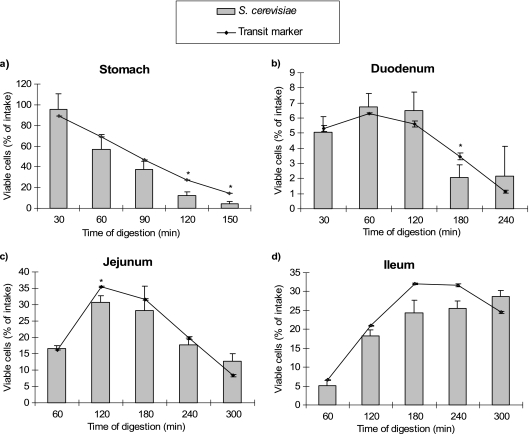
Our results also showed a high resistance of S. cerevisiae CNCM I-3856 to gastric and small-intestinal secretions and low gastric pH (Fig. 4). This result may encourage the use of this yeast in comparison to lactic acid bacteria in a probiotic strategy. Indeed, survival rates for Lactobacillus spp. and Streptococcus spp. during digestion in the TIM were much lower than those obtained in this study for yeasts (6, 21). In addition, our previous works on other S. cerevisiae strains showed the same yeast viability and also indicated that the survival rates obtained in vitro in TIM systems are consistent with in vivo data in humans (5).
FIG. 4.
Survival of S. cerevisiae CNCM I-3856 during in vitro digestions in the stomach (a), duodenum (b), jejunum (c), and ileum (d) of the TIM. The results obtained in each compartment for yeasts (gray bars) are compared with that of the transit marker, blue dextran (black line). Results are expressed as mean percentages ± SD (n = 4 digestions) of initial intake. *, S. cerevisiae results significantly different from those of the transit marker (P < 0.05).
In conclusion, our in vitro experiments provide new information on the survival of E. coli O157:H7 in the overall upper gastrointestinal tract of humans. Such data are essential for a full understanding of EHEC pathogenesis and for setting regulatory standards in the food processing industry. The S. cerevisiae CNCM I-3856 yeast strain appears to exert antagonistic effects against this enteric pathogen in the distal part of the small intestine, maybe through ethanol production. This property should be exploited for the development of prophylactic and/or therapeutic agents involved in the control of EHEC infections. The mode of action of S. cerevisiae CNCM I-3856, and particularly its interaction with the human intestinal microbiota, now needs to be further investigated using in vitro and in vivo approaches.
Acknowledgments
This work was supported by grants from the French Ministère de l'Education Nationale, de l'Enseignement Supérieur et de la Recherche to L.E.-M., ERT-CIDAM, and JE2526 USC-INRA 2018.
We thank Christine Martin (Unité de Microbiologie, INRA Clermont-Ferrand Theix) for the generous gift of the E. coli O157:H7 mutant. We also thank Lesaffre Company for providing the S. cerevisiae CNCM I-3856 strain. We are grateful to Sandrine Chalancon for technical assistance during the experiments on the TIM system.
Footnotes
Published ahead of print on 3 December 2010.
REFERENCES
- 1.Arnold, K. W., and C. W. Kaspar. 1995. Starvation- and stationary-phase-induced acid tolerance in Escherichia coli O157:H7. Appl. Environ. Microbiol. 61:2037-2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bach, S. J., T. A. McAllister, D. M. Veira, V. P. J. Gannon, and R. A. Holley. 2003. Effects of a Saccharomyces cerevisiae feed supplement on Escherichia coli O157:H7 in ruminal fluid in vitro. Anim. Feed Sci. Technol. 104:179-189. [Google Scholar]
- 3.Berger, C. N., et al. 2010. Fresh fruit and vegetables as vehicle for the transmission of human pathogens. Environ. Microbiol. 12:2385-2397. [DOI] [PubMed] [Google Scholar]
- 4.Bergholz, T. M., and T. S. Whittam. 2007. Variation in acid resistance among enterohaemorrhagic Escherichia coli in a simulated gastric environment. J. Appl. Microbiol. 102:352-362. [DOI] [PubMed] [Google Scholar]
- 5.Blanquet, S., J. P. Meunier, M. Minekus, S. Marol-Bonnin, and M. Alric. 2003. Recombinant Saccharomyces cerevisiae expressing P450 in artificial digestive systems: a model for biodetoxication in the human digestive environment. Appl. Environ. Microbiol. 69:2884-2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blanquet, S., et al. 2004. A dynamic artificial gastrointestinal system for studying the behavior of orally administered drug dosage forms under various physiological conditions. Pharm. Res. 21:585-591. [DOI] [PubMed] [Google Scholar]
- 7.Buts, J. P., and P. Bernasconi. 2005. Saccharomyces boulardii: basic science and clinical applications in gastroenterology. Gastroenterol. Clin. North Am. 34:515-532. [DOI] [PubMed] [Google Scholar]
- 8.Castagliuolo, I., J. T. LaMont, S. T. Nikulasson, and C. Pothoulakis. 1996. Saccharomyces boulardii protease inhibits Clostridium difficile toxin A effects in the rat ileum. Infect. Immun. 64:5225-5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiou, R. Y., R. D. Phillips, P. Zhao, M. P. Doyle, and L. R. Beuchat. 2004. Ethanol-mediated variations in cellular fatty acid composition and protein profiles of two genotypically different strains of Escherichia coli O157:H7. Appl. Environ. Microbiol. 70:2204-2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chong, Y., et al. 2007. Human intestinal tissue tropism in Escherichia coli O157:H7—initial colonization of terminal ileum and Peyer's patches and minimal colonic adhesion ex vivo. Microbiology 153:794-802. [DOI] [PubMed] [Google Scholar]
- 11.Czerucka, D., T. Piche, and P. Rampal. 2007. Review article: yeast as probiotics—Saccharomyces boulardii. Aliment. Pharmacol. Ther. 26:767-778. [DOI] [PubMed] [Google Scholar]
- 12.Dahan, S., et al. 2003. Saccharomyces boulardii interferes with enterohemorrhagic Escherichia coli-induced signaling pathways in T84 cells. Infect. Immun. 71:766-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Desreumaux, P., et al. 2010. Saccharomyces cerevisiae CNCM I-3856 reduces digestive discomfort and abdominal pain in subjects with irritable bowel syndrome: a randomized double-blinded placebo-controlled clinical trial, abstr. OP-181. Abstr. 18th United European Gastroenterology Week, Barcelona, Spain.
- 14.FAO/WHO. 2002. Report of a joint FAO/WHO working group on drafting guidelines for the evaluation of probiotics in food. Food and Agriculture Organization of the United Nations and World Health Organization, London, Ontario, Canada.
- 15.Fitzhenry, R. J., et al. 2002. Intimin type influences the site of human intestinal mucosal colonisation by enterohaemorrhagic Escherichia coli O157:H7. Gut 50:180-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Foligné, B., J. Dewulf, P. Vandekerckove, G. Pignède, and B. Pot. 2010. Probiotic yeasts: anti-inflammatory potential of various nonpathogenic strains in experimental colitis in mice. World J. Gastroenterol. 16:2134-2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Foster, J. W. 2004. Escherichia coli acid resistance: tales of an amateur acidophile. Nat. Rev. Microbiol. 2:898-907. [DOI] [PubMed] [Google Scholar]
- 18.Ganzle, M. G., C. Hertel, J. M. Van der Vossen, and W. P. Hammes. 1999. Effect of bacteriocin-producing lactobacilli on the survival of Escherichia coli and Listeria in a dynamic model of the stomach and the small intestine. Int. J. Food Microbiol. 48:21-35. [DOI] [PubMed] [Google Scholar]
- 19.Gobert, A. P., et al. 2007. Shiga toxin produced by enterohemorrhagic Escherichia coli inhibits PI3K/NF-kappaB signaling pathway in globotriaosylceramide-3-negative human intestinal epithelial cells. J. Immunol. 178:8168-8174. [DOI] [PubMed] [Google Scholar]
- 20.Mahajan, A., et al. 2009. An investigation of the expression and adhesin function of H7 flagella in the interaction of Escherichia coli O157:H7 with bovine intestinal epithelium. Cell. Microbiol. 11:121-137. [DOI] [PubMed] [Google Scholar]
- 21.Marteau, P., M. Minekus, R. Havenaar, and J. H. Huis in't Veld. 1997. Survival of lactic acid bacteria in a dynamic model of the stomach and small intestine: validation and the effects of bile. J. Dairy Sci. 80:1031-1037. [DOI] [PubMed] [Google Scholar]
- 22.Minekus, M., P. Marteau, R. Havenaar, and J. H. Huis in't Veild. 1995. A multicompartmental dynamic computer-controlled model simulating the stomach and small intestine. ATLA 23:197-209. [Google Scholar]
- 23.Ogawa, M., et al. 2001. Inhibition of in vitro growth of Shiga toxin-producing Escherichia coli O157:H7 by probiotic Lactobacillus strains due to production of lactic acid. Int. J. Food Microbiol. 68:135-140. [DOI] [PubMed] [Google Scholar]
- 24.Paton, J. C., and A. W. Paton. 1998. Pathogenesis and diagnosis of Shiga toxin-producing Escherichia coli infections. Clin. Microbiol. Rev. 11:450-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pradel, N., et al. 2000. Prevalence and characterization of Shiga toxin-producing Escherichia coli isolated from cattle, food, and children during a one-year prospective study in France. J. Clin. Microbiol. 38:1023-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rangel, J. M., P. H. Sparling, C. Crowe, P. M. Griffin, and D. L. Swerdlow. 2005. Epidemiology of Escherichia coli O157:H7 outbreaks, United States, 1982-2002. Emerg. Infect. Dis. 11:603-609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roering, A. M., et al. 1999. Comparative survival of Salmonella typhimurium DT 104, Listeria monocytogenes, and Escherichia coli O157:H7 in preservative-free apple cider and simulated gastric fluid. Int. J. Food Microbiol. 46:263-269. [DOI] [PubMed] [Google Scholar]
- 28.Roxas, J. L., et al. 2010. Enterohemorrhagic E. coli alters murine intestinal epithelial tight junction protein expression and barrier function in a Shiga toxin-independent manner. Lab. Invest. 90:1152-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takumi, K., R. de Jonge, and A. Havelaar. 2000. Modelling inactivation of Escherichia coli by low pH: application to passage through the stomach of young and elderly people. J. Appl. Microbiol. 89:935-943. [DOI] [PubMed] [Google Scholar]
- 30.Tamplin, M. L. 2005. Inactivation of Escherichia coli O157:H7 in simulated human gastric fluid. Appl. Environ. Microbiol. 71:320-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thanassi, D. G., L. W. Cheng, and H. Nikaido. 1997. Active efflux of bile salts by Escherichia coli. J. Bacteriol. 179:2512-2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tkalcic, S., et al. 2003. Fecal shedding of enterohemorrhagic Escherichia coli in weaned calves following treatment with probiotic Escherichia coli. J. Food Prot. 66:1184-1189. [DOI] [PubMed] [Google Scholar]
- 33.Wong, C. S., S. Jelacic, R. L. Habeeb, S. L. Watkins, and P. I. Tarr. 2000. The risk of the hemolytic-uremic syndrome after antibiotic treatment of Escherichia coli O157:H7 infections. N. Engl. J. Med. 342:1930-1936. [DOI] [PMC free article] [PubMed] [Google Scholar]