SUMMARY
Although both natural and induced regulatory T (nTreg and iTreg) cells can enforce tolerance, the mechanisms underlying their synergistic actions have not been established. We examined the functions of nTreg and iTreg cells by adoptive transfer immunotherapy of newborn Foxp3-deficient mice. As monotherapy, only nTreg cells prevented disease lethality, but did not suppress chronic inflammation and autoimmunity. Provision of Foxp3-sufficient conventional T cells with nTreg cells reconstituted the iTreg pool and established tolerance. In turn, acute depletion of iTreg cells in rescued mice resulted in weight loss and inflammation. Whereas the transcriptional signatures of nTreg and in vivo derived iTreg cells were closely matched, there was minimal overlap in their T cell receptor (TCR) repertoires. Thus, iTreg cells are an essential non-redundant regulatory subset that supplements nTreg cells, in part by expanding TCR diversity within regulatory responses.
INTRODUCTION
“Natural” regulatory T (nTreg) cells that express the forkhead or winged helix transcription factor Foxp3 arise in the thymus, where they require high affinity T cell receptor (TCR) ligation by an agonist peptide-MHC complex for Foxp3 induction (Jordan et al., 2001; Relland et al., 2009). “Induced” Foxp3+ regulatory T (iTreg) cells can be generated from naïve, mature CD4+ “conventional” T (Tconv) cells during T cell activation by both TGF-β dependent and independent mechanisms (Chen et al., 2003; Schallenberg et al.). The PD-L1-PD-1 ligand-receptor pathway and environmental factors, such as some aryl hydrocarbon receptor agonists and the vitamin A metabolite all-trans retinoic acid enhance iTreg cell conversion, suggesting that control of iTreg cell production is biologically important (Coombes et al., 2007; Francisco et al., 2009; Quintana et al., 2008). Collectively, Treg cells are required for the maintenance of immunological tolerance, as illustrated by the fatal autoimmune lymphoproliferative disease that develops in neonatal mice and humans deficient in Foxp3 (Brunkow et al., 2001; Chatila et al., 2000).
Both iTreg and nTreg cells have suppressive function, measured by their capacity to inhibit T cell proliferation and suppress experimental autoimmune disease by adoptive transfer immunotherapy (Fantini et al., 2006; Huter et al., 2008b; Mottet et al., 2003). However, the relative contribution of each cell type to the peripheral Treg cell compartment and to the maintenance of tolerance is largely unknown and may depend upon the particular model used to examine this question (Curotto de Lafaille and Lafaille, 2009; Haribhai et al., 2009).
The production of iTreg cells in vivo has been examined in several experimental systems. After adoptive transfer, polyclonal and TCR transgenic monoclonal populations of CD4+ T cells can be induced to express Foxp3, either during homeostatic expansion or by chronic intravenous antigen exposure (Apostolou and von Boehmer, 2004; Curotto de Lafaille et al., 2004). Oral administration of antigen also generates iTreg cells, and these contribute to antigen-specific tolerance in the gut (Mucida et al., 2005). When naïve CD4+ Foxp3− Tconv cells are transferred into Rag2−/− mice, the mice develop colitis, and approximately 10% of the CD4+ mesenteric lymph node cells induce Foxp3 expression. These in vivo derived iTreg cells contribute to successful adoptive transfer immunotherapy with nTreg cells and to tolerance in the intestinal tract (Haribhai et al., 2009). In contrast to these studies, peripheral conversion is not observed after an acute viral infection, and only 2–3% of proliferating TCR transgenic Tconv cells upregulate Foxp3 after immunization (Fontenot et al., 2005; Haribhai et al., 2007). TCR repertoire studies suggest anywhere from a 4–40% overlap between the Tconv and Treg TCR repertoires, depending upon the approach used for the study (Hsieh et al., 2004; Lathrop et al., 2008; Pacholczyk et al., 2006; Wong et al., 2007). Thus some contribution of iTreg cells to the peripheral Treg cell pool seems plausible, even in healthy animals.
Gene expression studies have compared the transcriptional signature of nTreg cells with that of iTreg cells produced in vivo and in vitro using a variety of conditions (Feuerer et al., 2010; Haribhai et al., 2009; Hill et al., 2007). These studies confirm that while iTreg cells share a fraction of the canonical nTreg cell transcriptosome, they also contain a large number of differentially enriched transcripts. This divergence is not attributable to T cell activation, but correlates with the method used to generate the iTreg cells. Maintenance of the iTreg transcriptional signature and the generation of suppressive function depend upon Foxp3 expression, analogous to the role of Foxp3 in nTreg cells (Haribhai et al., 2009; Hill et al., 2007). Methylation studies of conserved CpG motifs in the Foxp3 promoter also identify epigenetic differences between nTreg and iTreg cells, with the former showing more extensive demethylation and stable Foxp3 expression (Floess et al., 2007; Polansky et al., 2008). These data are consistent with the loss of Foxp3 expression in adoptive transfer studies using iTreg cells generated in vitro (Selvaraj and Geiger, 2007). Collectively, these genomic studies imply that peripheral Treg cells may be a complex mixture of sub-phenotypes able to interconvert (Feuerer et al., 2010). The transcriptional programs of stable iTreg cells derived in vivo and stable nTreg cells isolated under the same conditions from the same hosts have not been compared, and may provide additional insights into the mechanisms of iTreg cell function.
Although iTreg cells can clearly develop in vivo, the iTreg cell contribution to immunologic tolerance remains to be established. This question was first evaluated using neonatal adoptive transfer immunotherapy to treat C57BL/6 scurfy mice, which express a mutated and nonfunctional Foxp3 protein and develop a fatal autoimmune syndrome. Mice treated with either nTreg or iTreg cells alone showed a reduction in the pathological manifestations of the disease at the conclusion of the studies, 21 days after Treg cell transfers (Fontenot et al., 2003; Huter et al., 2008a). Tolerance at steady state in mature mice and the effect of combined therapy with both iTreg and nTreg cells were not considered in these studies.
In this report, we examined the respective roles of nTreg and iTreg cells by adoptive transfer immunotherapy of newborn Foxp3-deficient mice. Results demonstrated that in vivo-derived iTreg cells, which comprised ~15% of the peripheral Foxp3+ Treg cell pool, acted in concert with nTreg cells to enforce tolerance by mechanisms that involved expansion of the TCR repertoire of regulatory responses.
RESULTS
Rescue of Foxp3-deficient mice
The capacity of adoptively transferred Treg cell populations to rescue Foxp3-deficient mice is a comprehensive test of Treg cell function. We utilized BALB/c Foxp3K276X mice, which lack Foxp3 protein expression (Lin et al., 2005). Over 90% of BALB/c Foxp3K276X mice rapidly succumb to the autoimmune lymphoproliferative disease associated with Foxp3-deficiency within 21 days of birth, and no mice live beyond 32 days. Mice with the same mutation bred on to the C57BL/6 background live markedly longer, making it more difficult to evaluate the long-term effects of adoptive transfer immunotherapy in C57BL/6 mice (Figure 1A, BALB/c n=68, C57BL/6 n=19). Most subsequent experiments used BALB/c Thy1.2+ Foxp3K276X mice to provide a stringent therapeutic test with a clear endpoint in untreated mice.
Figure 1.
Adoptive transfer immunotherapy of newborn Foxp3-deficient mice. (A) Kaplan-Meier survival curves of Foxp3K276X mice bred onto the BALB/c (n=68) and C57BL/6 (n=19) backgrounds. (B) Kaplan-Meier survival curves of BALB/c Foxp3K276X mice rescued by neonatal intraperitoneal adoptive transfer with 60×106 unfractionated splenocytes (n=194) or purified 5×106 CD4+ T cells (n=45). (C) Representative flow cytometry analysis of lymphocytes from the indicated mice stained for naïve (CD62Lhi CD44lo) and effector or memory (CD62Llo CD44hi) CD4+ T cells. For these and all subsequent representative FACS plots, numbers indicate the mean percent of cells in the quadrant. Groups included untreated Foxp3K276X mice, n=4; Foxp3K276X mice rescued (R) with unfractionated splenocytes, n=4; 6–8 week old Foxp3EGFP control mice, n=16. The CD4+ Thy1.2+ gate identified host T cells in the rescued mice. (D) The serum concentration of pro-inflammatory cytokines from the indicated mice (n=10 for all groups). *p<0.05, **p<0.005, ***p<0.0005, ns-not significant.
To determine the number of Treg cells and the time course needed for successful therapy, we injected ~60×106 unfractionated syngenic splenocytes into the peritoneal cavity of newborn Foxp3K276X mice. These inocula contained approximately 0.9×106 nTreg cells and 5.8×106 Tconv cells. Most mice survived to 50 days (Figure 1B, 91 %, 177/194 mice), although survival decreased slightly at 100 days (86%, 166/194 mice). Surviving mice exhibited normal weight gain and were fertile (data not shown). Reduction of the inocula to 5×106 purified Thy1.1+ CD4+ T cells, containing approximately 0.7×106 nTreg cells, also resulted in effective rescue. Most mice survived to 50 days (89%, 40/45 mice), and survival decreased slightly by 100 days (82%, 37/45 mice). There was no statistical difference in survival between the two treatments at 50 days and at 100 days. Treated mice had normal percentages of naïve T cells and lacked the elevated serum concentrations of pro-inflammatory cytokines seen in Foxp3K276X mice (Figure 1C,D). Thus neonatal adoptive transfer immunotherapy with CD4+ T cells was sufficient to rescue Foxp3 deficient mice.
Monotherapy with nTreg or iTreg cells
Using these CD4+ T cell numbers as an estimate, we examined the capacity of different Thy1.1+ T cell populations to reconstitute Thy1.2+ Foxp3-deficient recipient mice. All transferred populations were purified by cell sorting from Foxp3EGFP mice, which contain a bicistronic Foxp3 locus that co-expresses Foxp3 and the enhanced green fluorescent protein (EGFP) under control of the endogenous Foxp3 promoter and enhancer elements (Haribhai et al., 2007). All mice were followed for at least 50 days to allow the development of partially treated phenotypes.
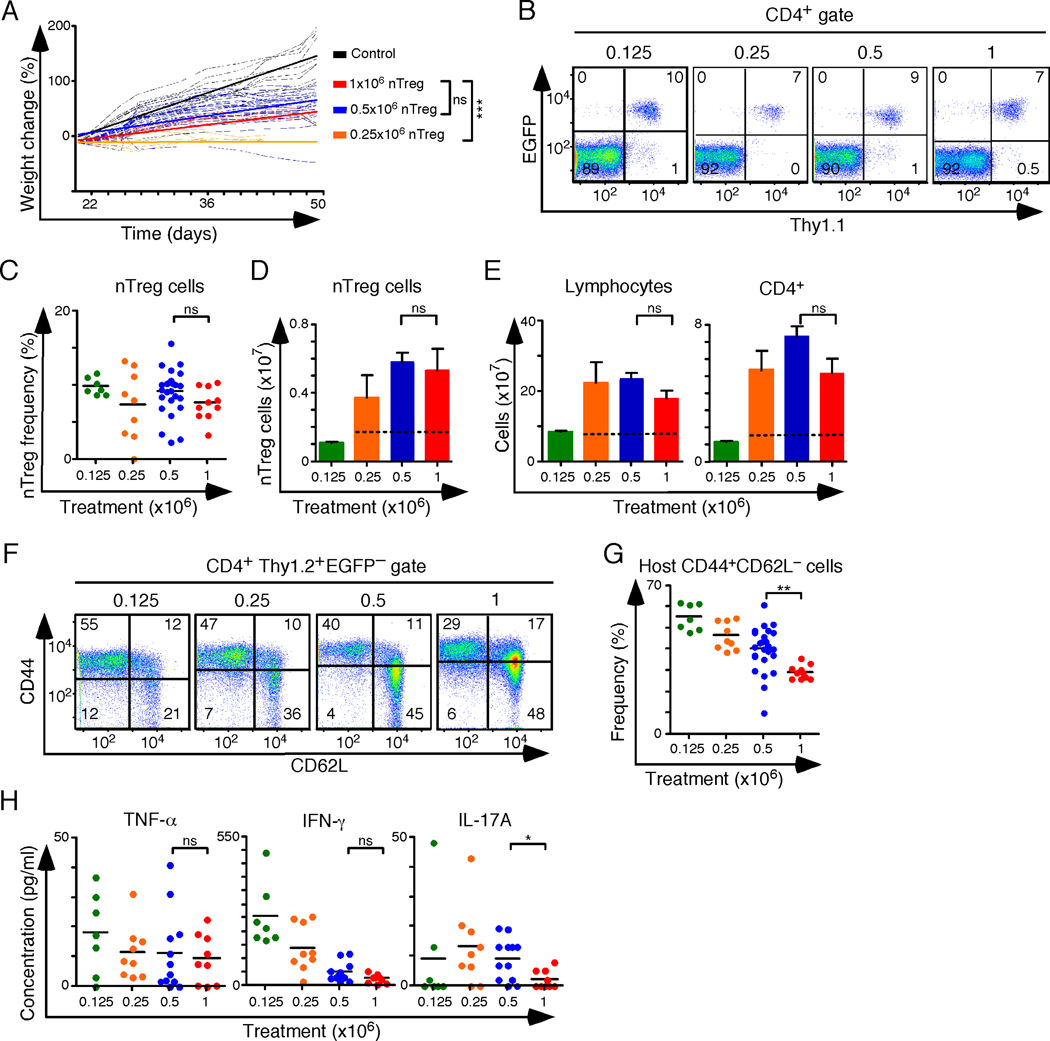
Neonatal transfer of 0.125×106 Thy1.1+ nTreg cells (CD4+EGFP+) isolated from Foxp3EGFP mice failed to prolong survival (n=16, survival data not shown). Mice lived longer but died after 30–45 days (n=9) when 0.25×106 Thy1.1+ nTreg cells were used for the rescue. Increasing the dose to 0.5×106 cells resulted in the survival of all mice to 50 days (n=26). Mice that received 0.5×106 nTreg cells gained weight, but at a reduced rate compared to controls (Figure 2A). Flow cytometry analysis of reconstituted mice showed 7–10% of CD4+ cells in the lymph nodes were EGFP+ nTreg cells, irrespective of the treatment group (Figure 2B, C). The average number of lymphocytes recovered from peripheral lymphoid tissues (spleens, brachial, axillary, superficial inguinal and mesenteric lymph nodes) of mice treated with 0.5×106 nTreg cells was significantly higher than that of untreated healthy (233±16×106 cells vs. 75±2.5×106 cells, p=0.0003) Similarly, the total number of CD4+ T cells (71±5×106 versus 15±1.6×106 cells, p=0.0003) and nTreg cells (6±0.6×106 versus 1.7±0.2×106 cells, p=0.0003) in this collection of lymph nodes and spleens was increased in the 0.5×106 nTreg cell treatment group relative to age-matched controls (Figure 2D, E). Treated mice also had a reduced frequency of naïve (CD62LhiCD44lo) host T cells (Figure 2F, G). These data established 0.5×106 nTreg cells as sufficient for survival to 50 days, although transfer recipients had splenomegaly, lymphoproliferation, and host T cell activation. Serum amounts of TNF-α, IFN-γ and IL-17A were also increased, suggesting incomplete tolerance (Figure 2H). Doubling the dose to 1.0×106 nTreg cells did not significantly increase weight gain, decrease most disease manifestations, or increase the number of nTreg cells recovered (Figure 2A–H). It therefore seemed unlikely that an additional increase in the nTreg cell dose above 1×106 cells would appreciably improve the therapeutic outcome. Furthermore, the 1.0×106 nTreg cell dose exceeded the average number of nTreg cells found in 60×106 unfractionated splenocytes, so we used 1.0×106 nTreg cells in all subsequent experiments. Replacing the nTreg cells with 1.0×106 in vitro generated iTreg cells had no significant effect on survival, and all mice died within 30 days (n=6, data not shown). In summary, nTreg cells survived and expanded in BALB/c Foxp3K276X mice, but were not sufficient to maintain tolerance. In vitro generated iTreg cells did not substitute for nTreg cells, at least when numerically equivalent cell doses were compared.
Figure 2.
The effect of nTreg cell monotherapy on therapeutic outcome. (A) Linear regression analysis of weight change over time following intraperitoneal adoptive transfer of the indicated number of nTreg cells into newborn Foxp3K276X mice. Mice treated with 0.125×106 nTreg cells did not survive to weaning and are not shown. (B) Representative flow cytometry analysis of lymph node cells gated on CD4+ T cells. Thy1.1 marks the transferred nTreg cell population. (C) Frequency of nTreg cells (CD4+ gate) in the lymph nodes of treated mice. (D, E) Bar graphs depicting the average sum (per mouse) of nTreg cells (D), unfractionated lymphocytes and CD4+ T cells (E) recovered from the spleen, axillary, brachial, superficial inguinal and mesenteric lymph nodes of treated mice and healthy 6–8 week old control mice. Where indicated, dashed horizontal lines mark the mean values for healthy 6–8 week old control mice (n=16). This control value does not apply to mice treated with 0.125 ×106 nTreg cells, which did not survive beyond 22 days. (F) Representative flow cytometry analysis of host CD4+ Tconv cells stained for CD44 and CD62L. (G) Frequency of CD44+CD62L− host T cells. (H) Serum concentration of TNF-α, IFN-γ, and IL-17A in treated mice. Colored dots represent individual mice and the horizontal bars indicate mean values. Treatment dose key: 0.125 ×106 nTreg cells (green, n=16); 0.25×106 nTreg cells (orange, n=9); 0.5×106 nTreg cells (blue, n= 27); 1.0×106 nTreg cells (red, n=10); control (black, n=9); 3–5 experiments per nTreg cell dose. ns - not significant, *p<0.05, **p<0.005, ***p<0.0005.
Therapy with iTreg and nTreg cell combinations
In Foxp3K276X mice treated with nTreg cells alone, all host CD4+ Tconv cells carried a nonfunctional Foxp3 allele and could not become iTreg cells. In the experiments with either unfractionated splenocytes or purified CD4+ T cells (Figure 1), CD4+ Tconv cells might have given rise to iTreg cells that acted synergistically with nTreg cells to promote tolerance, as we have previously shown in an experimental model of colitis (Haribhai et al., 2009). To further evaluate the contribution of iTreg cells to tolerance, we combined nTreg cells with Tconv cells in the rescue experiments. Adoptive transfer of 1×106 Thy1.2+ nTreg cells together with 4×106 Thy1.1+ Tconv cells resulted in the survival of 28/29 transfer recipients and in improved weight gain (Figure 3A). At 50 days, Thy1.1+ in vivo derived iTreg cells comprised about 1% of CD4+ T cells, 15% of the total EGFP+ Treg cell pool, and about 1/3 of the surviving Thy1.1+ cells (Figure 3B, C). The frequency of nTreg cells was similar between the treatment groups (Figure 3C), while the total number of Treg (iTreg + nTreg) cells recovered from the spleens and peripheral lymph nodes of treated mice was decreased from 5.2×106 to 2.8×106 cells (Figure 3D). Antigen-specific Treg cells may expand in some diseases that are associated with chronic inflammation, thus a reduction in Treg cell numbers can be consistent with a sustained decrease in disease activity (Curotto de Lafaille et al., 2008; Miyara et al., 2006; Schmitz-Winnenthal et al., 2009). Mice that received 1.0×106 nTreg cells plus 4×106 Tconv cells also showed reductions in several of the disease manifestations associated with Foxp3 deficiency. The total numbers of lymphocytes (data not shown), CD4+ T cells and B cells were reduced to numbers similar to those of normal mice (Figure 3E). The proportion of naïve host T cells increased from 48% to 56% (Figure 3F, G).
Figure 3.
In vivo derived iTreg cells contribute to tolerance. (A) Linear regression analysis of weight change over time following intraperitoneal adoptive transfer of the indicated number of nTreg and Tconv cell populations into newborn Foxp3K276X mice (red − 1.0×106 nTreg cells, n=10; green − 1×106 nTreg cells + 4×106 Tconv cells, n=29; orange − 1×106 nTreg cells + 4×106 ΔTconv cells from Foxp3ΔEGFP mice, n=8; blue − 1×106 nTreg cells + 1×106 iTreg cells generated in vitro, n=6; 3–7 experiments per treatment group. (B) Representative FACS analysis of lymph node cells from the indicated groups. Analysis is gated on CD4+ T cells. Thy1.1 marks different transferred populations: nTreg cells in 1st and 3rd panels, Tconv cells in the 2nd panel, or in vitro produced iTreg cells in the 4th panel. (C) Frequency of nTreg and iTreg cells. (D, E) Bar graphs depicting the average sum of nTreg cells (D), CD4+ T cells or CD19+ B cells (E) recovered from the spleen, axillary, brachial, superficial inguinal and mesenteric lymph nodes of treated mice. Dashed horizontal lines mark the mean values for healthy 6–8 week old control mice (n=16), and iTreg cells are shown in pink. (F) Representative FACS analysis of host CD4+ Tconv cells stained for CD44 and CD62L. (G) Frequency of CD44+CD62L− host T cells. (H) Representative sections from the colons of the indicated mice stained with hematoxylin and eosin. (I) Colitis scores from treated mice. Colored dots represent individual mice and the horizontal bars indicate mean values. (J) Serum concentration of TNF-α, IFN-γ, and IL-17A in treated mice. Data from 1×106 nTreg cell treatment group shown in Figure 2 are repeated here to facilitate direct comparison. *p<0.05, **p<0.005, ***p<0.0005.
Inflammatory bowel disease is a prominent feature of Foxp3 deficiency. However, mice treated with nTreg cells alone still developed transmural lymphocytic infiltration in the colon. While extensive, this infiltrate was not associated with a marked reduction in mucin production or with extensive crypt elongation (Figure 3H). When the Treg cell compartment of treated mice contained both nTreg and iTreg cells, the infiltrates were reduced, as reflected by decreased colitis scores (Figure 3I). Tissue infiltration of other organs, including the lung and liver, were also reduced (data not shown) and serum concentrations of TNF-α, IFN-γ, and IL-17A were lower, consistent with improvement in the other disease manifestations (Figure 3J). Thus a stable population of iTreg cells was produced in vivo from Tconv cells and this process correlated with marked reductions in lymphoproliferation, cytokine production, and pathologic tissue infiltration of many organs, including the colon.
To ascertain that the contribution of CD4+ Tconv cells to the rescue was Foxp3-dependent, we used rescued Thy1.1+ Foxp3ΔEGFP mice as the source of naïve Thy1.1+ ΔTconv cells. Foxp3ΔEGFP mice express a nonfunctional Foxp3ΔEGFP fusion protein and untreated mice develop an autoimmune lymphoproliferative disorder that is indistinguishable from that of Foxp3K276X mice (Lin et al., 2007). We therefore combined 1×106 Thy1.2+ nTreg cells from Foxp3EGFP mice with 4×106 Thy1.1+ ΔTconv cells from rescued Foxp3ΔEGFP mice and used these cells in our treatment experiments. Linear regression analysis of the weight gain, where the ΔTconv cells lacked a functional Foxp3 allele, overlapped that seen when nTreg cells were used alone and was reduced when compared to the nTreg + Tconv cell transfer experiments (Figure 3A, n=8). Other manifestations of disease were worse and included increased CD4+ T cell activation and proliferation, B cell proliferation, and elevated serum concentrations of pro-inflammatory cytokines (Figure 3B–J). These measurements indicated a level of disease activity that was similar to that seen when nTreg cells were used alone, confirming that it was the capacity of Tconv cells to express Foxp3 that was associated with their therapeutic potential.
Next, we determined the effect of reducing the number of iTreg cells generated in vivo by transferring 1×106 nTreg plus 1×106 Tconv cells, which decreased the number of Tconv by 75%. The data showed incomplete rescue, with host T cell activation, tissue infiltration and colitis scores that fell between monotherapy with nTreg cells alone and the combination therapy with 1×106 nTreg plus 4×106 Tconv cells (Figure S1). Although the total number of Treg cells present at the end of the experiments was similar in the two treatment groups, the frequency and number of iTreg cells reflected the size of the starting Tconv cell population. Similar results were obtained when we replaced the missing Tconv cells with 3×106 EGFP− DO11.10 TCR transgenic T cells (Figure S2). At 50 days, approximately 3.6% of CD4+ T cells expressed the DO11.10 TCR, and none of these were EGFP+. These data demonstrated that a reduction in iTreg cell numbers reduced their clinical effect, that the frequency and number of iTreg cells reflected the size of the starting Tconv cell population, and that iTreg cells are generated and maintained through antigen specific selective pressure.
Last, we replaced the Tconv cells with iTreg cells produced in vitro, as this approach is a feasible source of Treg cells when considering adoptive transfer immunotherapy in humans. It seemed plausible that those iTreg cells generated in vivo in response to the specific needs of the host might be functionally different than the iTreg cells derived in vitro from a randomly selected polyclonal population. To compare the two types of iTreg cells, we treated Foxp3-deficient pups with 1×106 in vitro generated Thy1.1+ iTreg cells and 1×106 Thy1.2+ nTreg cells. All mice that received a combination of in vitro generated iTreg cells and nTreg cells survived and their weight gain exceeded that seen when nTreg cells were used alone (Figure 3A, n=6). Approximately 50% of the surviving iTreg cells remained EGFP+ and comprised 4% of the total EGFP+ Treg cell population, consistent with long-term stability of iTreg cells generated in vitro (Figure 3B). Markers of disease activity improved relative to mice that received nTreg cells alone, and were similar to the transfers where mice received nTreg cells plus the higher dose of Tconv cells (Figure 3C–J). Thus, in vitro generated iTreg cells could largely substitute for iTreg cells derived in vivo, indicative of the functional similarity of the two iTreg cell populations.
Depletion of iTreg cells
Maintaining tolerance requires the continued presence of nTreg cells, as determined by nTreg cell depletion studies in adult mice (Kim et al., 2007). To investigate the role of iTreg cells in maintaining tolerance, we utilized a similar strategy involving selective iTreg cell depletion from successfully treated mice. For these experiments, we created C57BL/6 transgenic mice that expressed the diphtheria toxin receptor (DTR) under the control of the Foxp3 promoter (Foxp3DTR mice, Figure S3). In Foxp3DTR mice, a single 50µg/kg injection of diphtheria toxin (DT) resulted in elimination of 90% of peripheral blood nTreg cells by 24 hours. In vitro conversion experiments demonstrated that 94% of Foxp3+ iTreg cells expressed the DTR. We next bred Foxp3DTR mice to CD45.1+ Foxp3EGFP mice and used their progeny as a source of marked CD4+ Tconv cells capable of expressing both EGFP and the DTR upon appropriate stimulation in vivo.
Following the protocol used for the experiments in Figure 3, we treated newborn CD45.2+ C57BL/6 Foxp3ΔEGFP mice with 1×106 CD45.2+ EGFP+ nTreg cells mixed with 4×106 CD45.1+ EGFP−DTR− Tconv cells. Littermate controls received 1×106 CD45.2+ EGFP+ nTreg cells mixed with 4×106 CD45.1+ EGFP− Tconv cells that lacked the DTR transgene. Both groups of mice gained weight up to 50 days, similar to the previous treatment experiments performed in the BALB/c background. At 50 days, all mice were injected with 50 µg/kg of DT every other day for 15 days and analyzed 5 days later. Only those mice where the transferred Tconv cells carried the DTR transgene lost weight (Figure 4A). Flow cytometry analysis of the spleens and lymph nodes from control mice revealed that half of the surviving CD45.1+ cells expressed Foxp3, and that 33% of all EGFP+ Treg cells were iTreg cells derived by conversion from Tconv cells. In mice that received Tconv cells carrying the DTR transgene, there were no EGFP+CD45.1+ iTreg cells remaining after the series of injections, although the population of CD45.1+ EGFP− T cells was maintained (Figure 4B). While the frequency and number of nTreg cells in both groups was similar (Figure 4C,D), the frequency of host CD4+ T cells expressing the FoxpΔEGFP allele (dim EGFP signal) increased from 5% to 9% in iTreg cell-depleted mice (Figure 4B, middle boxed cells). This suggested pressure to replace the depleted iTreg cells. Lymphoproliferation and an increase in the frequency of host CD44hi CD62Llo CD4+ T cells occurred following iTreg cell depletion, and a higher proportion of host CD4+ T cells expressed IFN-γ (Figure 4E–I). The lungs from iTreg cell-depleted mice showed increased perivascular and peribronchiolar inflammatory infiltrates that contained macrophages, plasma cells, lymphocytes and mononuclear cells (Figure 4J). In addition, there were prominent interstitial infiltrates, with mononuclear cells and macrophages in the alveolar spaces. The liver, small intestine and colons also contained mixed inflammatory infiltrates (Figure 4K). These data demonstrated a pathologic increase in host T cell activation in the absence of iTreg cells that was most marked at mucosal surfaces and was consistent with a requirement for iTreg cells throughout the lifespan of the mouse.
Figure 4.
Selective iTreg cell depletion from treated C57BL/6 Foxp3ΔEGFP mice. (A) Weight change over time following intraperitoneal adoptive transfer of 1×106 Foxp3EGFP nTreg cells + 4×106 Foxp3EGFP Tconv cells (blue, n=3) or 1×106 Foxp3EGFP nTreg cells + 4×106 Foxp3EGFP/DTR+ Tconv cells (red, n=3) into newborn Foxp3-deficient mice. Diphtheria toxin was injected daily for 15 days beginning at 50 days of age (arrow) and analyzed 5 days later. Two experiments were performed. (B) Representative flow cytometric analysis of lymph node cells gated on CD4+ T cells. CD45.1 marks the transferred Foxp3EGFP and Foxp3EGFP/DTR+ Tconv cell populations. (C) Frequency of nTreg and iTreg cells following DT treatment. (D, E) Average numbers (±S.E.M.) of nTreg cells (D), total lymphocytes and CD4+ T cells (E) isolated from peripheral lymphoid tissues of Foxp3EGFP and Foxp3EGFP/DTR+ mice post DT treatment. (F, G) FACS analysis of CD44 and CD62L expression (F) and frequency of CD44+CD62L− cells (G) in host (Foxp3ΔEGFP) CD4+ Tconv. (H, I) Representative FACS analysis (H) and cell frequencies (I) of host CD4+ Tconv cells stained for IFNγ pre- and post-DT treatment. (J–K) Representative sections from the lung (J), and colon, liver, and small intestine (K) of the indicated mice stained with hematoxylin and eosin.
Phenotypic and transcriptional analysis of Treg cell subsets
We investigated the molecular basis for the contribution of in vivo derived iTreg cells to tolerance by comparing their cell surface phenotype and gene expression profile with that of nTreg cells from the same mice. Flow cytometric analysis of iTreg and nTreg cells from Foxp3K276X mice treated with 1×106 nTreg cells and 4×106 Tconv cells showed that both Treg cell subsets had similar cell surface levels CD25, CD62L, CD44, and CD103, GITR, Helios, and CTLA-4 (Figure 5A). Expression of Programmed death-1 (PD-1), which plays an important role in iTreg formation and function, was increased on iTreg cells (Francisco et al., 2009). Granzyme B was increased in nTreg cells, and iTreg cells had a minor population that did not express Helios. Thus we could only identify a few subtle differences in phenotype based on expression of several cell surface and intracellular proteins.
Figure 5.
Comparison of iTreg and nTreg cells from treated mice. (A) Representative cell surface marker analysis of CD4+EGFP+ lymph node iTreg (red) or nTreg (blue) cells stained as indicated. (B) Hierarchical clustering of probe sets significantly regulated in iTreg cells derived in vivo (top), n Treg cells from the same mice (middle), and in vitro derived iTreg cells 72 hours after Foxp3 induction (bottom), versus naïve Tconv cells from healthy mice. The scale represents fold change relative to Tconv cells (−3 fold to +3 fold). (C) Venn diagram depicting the number of commonly and uniquely regulated probe sets in the in vivo derived iTreg and nTreg cell transcriptional signatures. Bar graphs compare the fold change in expression of select prototypical genes found within the canonical Treg cell transcriptosome. (D) Venn diagram depicting the number of commonly and uniquely regulated probe sets in the in vivo derived iTreg and in vitro derived iTreg cell genetic signatures. Bar graphs compare the fold change in expression of the same genes shown in Figure 6c. (E) Methylation status of the CpG motifs in the Treg cell-specific demethylation region (TSDR) of conserved non-coding sequence 2 (CNS2) in Foxp3. Residue numbers represent individual CpG motifs in reference to the transcription initiation site of Foxp3.
Next, we compared the gene expression profile of pooled EGFP+ Thy1.1+ iTreg cells with that of EGFP+ Thy1.1− nTreg cells sorted from the spleens and lymph nodes of treated mice (Figure 5B). In aggregate, there were 3491 probe sets that were differentially expressed relative to naïve Tconv cells. Of these, 2651 probe sets (76%) were common to both iTreg and nTreg cells, with 1290 that were overexpressed and 1361 that were underexpressed (Figure 5C, Table S1). Overexpressed genes that are prototypical of the nTreg cell transcriptosome included Itgae, Izkf2, Ctla4, Il2ra, Nrp1, Il10, Dusp4, Icos, Socs2, Ahr, and Ikzf4. Underexpressed genes included Cd4, Itk, Ikzf1, Sox4, Il4ra, Il6ra, and Il7r. This set of common probe sets contained about half (302/603) of the microarray probes that defined the Treg cell genetic signature in another study (Hill et al., 2007). A direct comparison of probe sets within the intersection identified only 4 genes with significant differences in expression between the two types of Treg cells (Eif2s3y, Ddx3y, Kdm5d and C1qb). Further analysis showed that nearly all probe sets identified as “unique” were directionally concordant, being over expressed or repressed in both iTreg and nTreg cells relative to naïve Tconv cells, albeit at different levels (Tables S2 and S3). Another 10% (62/603) of the canonical Treg cell probe sets were found among these groups. Thus the gene expression profiles of iTreg and nTreg cells isolated from treated mice were similar to each other and to the published nTreg cell genetic signatures.
The gene expression profile of iTreg cells produced in vitro and analyzed 3 days after induction is distinct from that of nTreg cells isolated from healthy animals (Haribhai et al., 2009). Other data demonstrates that there are important transcriptional differences between iTreg cell subsets based on the method of Foxp3 induction and the stability of the corresponding iTreg cell phenotype (Feuerer et al., 2010). However, when we compared the genetic signatures of iTreg cells established in vivo with newly generated iTreg cells produced in vitro, we found striking differences in the gene expression profiles. A total of 5915 probe sets were differentially regulated when compared to Tconv cells. The intersection between these two iTreg cell subsets contained only 1150 probe sets (Tables S4–S6, Figure 5D). A number of prototypical Treg cell genes were not identified as commonly regulated, including Ikzf2, Klrg1, Il1rl1, S100a8, CD83, Vcam1, Gzma, Klf4, Tnfsf4, Tlr4, Il10, and Bcl10. The iTreg cell intersection included 37% of the nTreg cell transcriptosome identified by Hill et al (Hill et al., 2007). Thus iTreg cells produced and maintained in vivo are far more similar to nTreg cells from the same mice than to iTreg cells generated in vitro. While in vitro derived iTreg cells are clearly functional, they may require additional signals to acquire genetic and phenotypic stability. Interestingly, CpG islands in the Foxp3 promoter of in vivo derived iTreg cells were extensively methylated, suggesting that Foxp3 expression in iTreg cells is either less stable than that of nTreg cells or is maintained by factors other than this type of epigenetic mechanism (Figure 5E).
TCR repertoire comparisons of Treg cell subsets
The similar transcriptional profiles of nTreg and iTreg cells implied common mechanisms of suppression. Other factors, such as differences in antigenic specificity, might therefore underlie the requirement for iTreg cells and the selective pressure to retain iTreg cells in treated mice. To investigate this possibility, we isolated iTreg cells derived from Tconv cells and nTreg cells from spleens and lymph nodes (brachial, axillary, mesenteric, and superficial inguinal) of three treated BALB/c mice and compared their TCR repertoires by TCRβ spectratype analysis and CDR3 sequencing. Each treated mouse received a different aliquot of cells pooled from 2–3 mice as in previous experiments. Fragment analysis using 22 Vβ-Cβ primer pairs showed a predominantly Gaussian length distribution for most Vβ in the nTreg cell populations with modest skewing away from this pattern in iTreg cells (Figure S4). These data indicated that a broad distribution of β-chains was found in both nTreg and iTreg TCRs.
To compare the TCR repertoires of iTreg and nTreg cells, we selected Vβ2 and Vβ8.3 for further analysis based the similar distribution of CDR3 lengths seen in all 3 mice (Figures 6A, Figure S4). CDR3 regions from each mouse were amplified by PCR, cloned and sequenced, and the sequences compared at both the nucleotide and amino acid levels. We obtained a combined total of 3289 in-frame reads from 1509 iTreg and 1780 nTreg clones, and these encoded 1964 unique CDR3 polypeptides. (Table S7, Figure S5). Nucleotide sequence analysis revealed that the pattern and frequency of Jβ utilization was associated with the Vβ segment and not with the type of Treg cell (Figure 6B). CDR3 amino acid sequence analysis for each Vβ revealed that there was limited overlap between the predicted iTreg and nTreg CDR3 polypeptides (Figure 6C, 3–15% overlap). In agreement with this observation, the Morista-Horn similarity index (MHI) was <0.1 in 3 of the 6 iTreg versus nTreg TCR CDR3 comparisons, indicating very little similarity between the two populations (Figure 6C). Ln-rank versus ln-rank frequency plots revealed an excellent fit for a power law-like component in 10/12 of the repertoires analyzed (R2 > 0.95). Because of this power law-like relationship, the distributions are similar at all scales of measurement. Several repertoires also contained a second component consisting of a few high rank clonotypes, as seen in the analysis of recall responses (Naumov et al., 2006).
Figure 6.
TCR repertoire analysis of iTreg and nTreg cells from treated mice. (A) Representative iTreg and nTreg cell spectratype analysis using Vβ2 or Vβ8.3 5’ PCR primers. (B) Bar graphs comparing the utilization of Jβ segments by Vβ2 and Vβ8.3-containing iTreg and nTreg cell clones. (C) Venn diagrams showing the distribution of unique and overlapping iTreg and nTreg cell CDR3 amino acid sequences of Vβ2 and Vβ8.3-containing clones isolated from individual mice. The number of sequences in each category and the Morsita-Horn Similarity Index (MHI) for the comparison are shown. Ln-rank versus ln-rank frequency plots for each repertoire are shown below the corresponding Venn diagram.
Consistent with the Tconv cell origin of iTreg cells, MHI values from pooled Vβ sequence comparisons (Figure S6) were similar to those from other studies that compared the TCRα repertoires of nTreg cells with that of Tconv cells in mice expressing a transgenic TCRβ-chain (Hsieh et al., 2006; Pacholczyk et al., 2006; Wong et al., 2007). Most non-overlapping sequences were found in a single individual, confirming that Treg cell TCR repertoires are largely private (“private” TCRs, average 97.5%). For the handful of overlapping sequences, the situation was reversed and many CDR3 sequences were recovered from 2 or more individuals (“public” TCRs, average 58%). Most of the frequently recovered (≥4 times) clones were found among the overlapping sequences and many of these were generated by more than one DNA rearrangement (convergent sequences) involving the addition of “N region” nucleotides not found in the germline.
Among Vβ8.3 sequences, two CDR3 regions were seen 3–4 times more frequently than the next most common CDR3 regions. Clone 1 from mouse 3 had a CDR3 length of 14 amino acids (CASSDLGHLNTEVFF) and was recovered 29 times from the iTreg cell library and 26 times from the nTreg cell library, indicating this favored TCRβ chain was selected into both nTreg and Tconv cell lineages. This unusual occurrence generated a marked increase in the MHI relative to the percent overlap. Removing this one sequence from the analysis resulted in an MHI of 0.148, a value in line with the other comparisons. In contrast to this overlapping sequence, clone 56 from mouse 2 had a CDR3 length of 13 amino acids (CASRETGGYAEQFF) and was recovered 42 times from the nTreg cell library only. In aggregate, these data demonstrated that the iTreg and nTreg TCR repertoires were largely “private” and non-overlapping. “Public” β-chains occurred rarely, although when present they were 20-times more likely to be found in TCRs that contributed to both iTreg and nTreg responses.
DISCUSSION
Foxp3-deficient mice have an otherwise intact immune system, and they lack systemic inflammation in the early neonatal period. These features of Foxp3 deficiency, together with the aggressive nature of the autoimmune disease that subsequently develops in untreated mice, made this an ideal model for our studies designed to examine the role of in vivo-derived iTreg cells in tolerance induction. The studies herein identify iTreg cells as an essential, non-redundant regulatory subset that acts synergistically with nTreg cells to enforce peripheral tolerance.
Although nTreg cells were provided by adoptive transfer immunotherapy shortly after birth, we found that the frequency of in vivo derived iTreg cells was substantial, as it was in the lymphopenia-colitis model (Haribhai et al., 2009). We also found that iTreg cells generated in vitro through a TGFβ-dependent pathway largely substituted for those iTreg cells derived in vivo, suggesting that the two sources of iTreg cells were functionally equivalent. We did not directly examine the TGFβ dependence of the in vivo production of iTreg cells in this model, so a significant contribution from the TGFβ-independent CD25+CD69highCD62LintFoxp3− Treg cell precursor population cannot be excluded (Schallenberg et al., 2010). Indeed, the recovery of half as many iTreg cells after 50 days, when the iTreg cells were derived in vitro rather than in vivo, might argue that more than one iTreg production pathway is operational. Regardless of the mechanisms supporting iTreg cell induction, these data reinforce the conclusion that the iTreg cell induction pathways are important tolerogenic mechanisms, both in the setting of chronic inflammation and in normal individuals.
Our initial investigations into the mechanisms underpinning the functional synergy between iTreg and nTreg cells focused on comparing the gene expression profiles of the two cell types, which were found to be remarkably similar. One explanation for this similarity is that each transcriptional signature is a composite of several Treg cell subsets, which could tend to blur distinctions between the two Treg cell types (Feuerer et al.). Nevertheless, the current data argue strongly that iTreg and nTreg cells share overlapping effector mechanisms. Gene expression profiles do not exclude a skewed reliance on certain Treg cell mediators like IL-10, which showed 2.2 fold increase in the in vivo derived iTreg cells over nTreg cells isolated from the same mice. Other potential distinctions between the two Treg cell subsets include increased expression of PD-1 by iTreg cells and Gzm B by nTreg cells, and differences in localization. We also found that the transcriptional signature of iTreg cells produced in vivo was more similar to nTreg cells than to iTreg cells produced in vitro. The most likely explanation for the differences in gene expression between iTreg cells produced in vivo and in vitro is that the profile of iTreg cells generated in vitro is largely a function of TCR, TGFβ and IL-2-mediated signaling, at least shortly after induction (Hill et al., 2007). Indeed, the gene expression profile of iTreg cells produced in vitro 72 hours after induction is independent of Foxp3 (Haribhai et al., 2009). We have not yet examined the transcriptome of iTreg cells produced in vitro and subsequently allowed to achieve equilibrium in treated mice. Based on the current results, this gene expression profile is anticipated to closely match that of both iTreg cells derived in vivo and nTreg cells.
If iTreg and nTreg cells share similar effector mechanisms, as suggested by their overlapping gene expression signatures, then other factors must contribute to the capacity of iTreg cells to complement the function of nTreg cells. For example, a synergistic interaction could develop if iTreg cells provided different TCR specificities than those found in nTreg cells, as we observed. These results shed light on the origins of in vivo-derived iTreg cells. Published data demonstrates that some nTreg cells can lose Foxp3 expression and survive as ” ex-Treg” cells (Yang et al., 2008; Zhou et al., 2009). In our experiments, such cells would be sorted with the Tconv cells and might re-express Foxp3 after transfer (Lathrop et al., 2008). A substantial overlap between nTreg and iTreg cells could therefore indicate that the two populations were clonally related, implying that iTreg cells were “ex-Treg” cells that reacquired Foxp3 expression and suppressive capability under the conditions of our experiment. Similarly, iTreg cells could be a special population of Tconv cells poised to express Foxp3 based on their shared TCR specificity with nTreg cells (Schallenberg et al., 2010). Again, substantial TCR repertoire overlap would be predicted if this shared specificity was created at the clonal level during the cellular expansion that occurs following β-selection in the thymus. From another perspective, iTreg cells might come from those Tconv cells that bear no clonal relationship to nTreg cells, based on the well-documented differences in the TCR ligand affinity requirements for the selection of nTreg and Tconv cells in the thymus (Jordan et al., 2001). Here, the predicted result would be essentially no TCRβ CDR3 overlap between the two populations, which is largely what we observed, particularly when considering that peripheral Treg cells probably contain a mixture of nTreg and iTreg cells. A recent estimate places the frequency of iTreg cells in the peripheral Treg compartment at 4–7% (Lathrop et al., 2008). We estimate that the iTreg cell frequency in rescued Foxp3 mutant mice is closer to 10–15%. Such misidentified iTreg cell TCRs would create an apparent overlap in the iTreg versus nTreg cell TCR comparisons and may well be a contributor to the small overlap observed in our studies. Thus our work is most consistent with naturally arising nTreg and iTreg cell populations with distinct TCR repertoires, indicating the two populations are clonally unrelated.
Since there are theoretically 1015 different TCRs that can be generated, the <1×106 different TCRβ chains found within an individual mouse virtually assures that these randomly selected repertoires are largely unique to each individual (i.e. “private”) (Quigley et al., 2010). Thus we expected that there would be little overlap in TCRβ CDR3 sequences from mouse to mouse, as we observed. However, a much higher frequency of the overlapping sequences in our experiments are “public”, which could indicate that these were the most abundant clones in the donor mice, that iTreg and nTreg cells share clonal identity, and that the repertoires are largely overlapping. Several lines of evidence argue against this point of view. First, many private nTreg and iTreg sequences were equally abundant, based on the frequency of their recovery. Thus abundant clones are not restricted to the overlapping component of the repertoires. Second, if one source of overlap derives from misidentified iTreg cells within the transferred nTreg pool, these clones may be numerically advantaged at the time of transfer, relative to new iTreg cells that arise in transfer recipients. Furthermore, misidentified iTreg clones that are present in the nTreg cell inoculate may be particularly important, since they arise in healthy mice. It follows that these clones would be found frequently in the overlapping component of the Treg repertoires, and that they would also be more likely to be public if they fill a ubiquitous antigen-specific niche left vacant by nTreg cells. Indeed, public sequences are seen in well-characterized antigen-specific CD4+ and CD8+ Tconv cell responses (Kedzierska et al., 2004; Menezes et al., 2007).
A different explanation for the overlapping “public” CDR3 sequences deserves special attention, because it has little to do with antigen specificity. It has been proposed that an essential characteristic of “public” CDR3 sequences is that they are generated by “convergent recombination” (Venturi et al., 2008). In this proposal, the number of different ways that a particular TCR CDR3 amino acid sequence can be generated by germline recombination is an important determinant of TCR production frequency and therefore of TCR sharing, quite apart from the antigen specificity of the TCR (Quigley et al., 2010). In other words, certain TCRβ amino acid sequences are more likely to be “public” because they are more frequently generated in the thymus. In our studies, public TCRβ sequences generated by convergent recombination would also be more likely to be overlapping, since each new TCRβ sequence is paired with a different TCRα chain, generating a new TCR and an independent opportunity for clones to be selected into either the iTreg or nTreg pool. Indeed, we found that the most frequently identified public, overlapping TCRβ chain amino acid sequence was generated by 7 unique nucleotide sequences. Some of our data therefore supports this latter model.
It is important to note that in our experiments, the size and complexity of both Treg cell TCR repertoires is fixed, since there can be no new production of Foxp3+ cells by the host. This experimental design may have expanded the opportunity to observe the generation of iTreg cells in vivo as well as the expansion of a few high rank clonotypes. Most of our TCR repertoires fit a two-component model, with a power law-like component and a component consisting of a few high rank clonotypes. This indicates that peripheral selection events occur after adoptive transfer. Given the repertoire differences described herein, coupled with the profound differences in the thymic selection requirements of nTreg and Tconv cells, it seems very unlikely that iTreg and nTreg cells will recognize identical sets of self-antigens and foreign antigens and that they will do so at comparable frequencies. Nevertheless, we make no a priori claim about the antigen specificity of the responses analyzed, and the reader should draw a distinction between diversity (unique TCR sequences) and specificity (antigens recognized).
In rescued mice that received both nTreg and either Thy1.1+ Tconv or Thy1.1+ in vitro generated iTreg cells, a substantial number of Thy1.1+ Foxp3− cells were maintained long-term. There are a few possibilities for the origin of these cells. A bidirectional linear model where Tconv cells gain and lose Foxp3 expression based on the needs of the host has been proposed and is consistent with the data (Haribhai et al., 2009). In a variation of this view, some Treg cells are not terminally differentiated and can adopt alternative TH cell fates (Zhou et al., 2009), or co-express other TH lineage specification factors that alter their distribution and homeostatic properties (Koch et al., 2009). It will be important to determine the cytokines produced by these Thy1.1+ Foxp3− cells, to compare their TCR repertoire with that of iTreg and nTreg cells, and to use genetic tools to map cell fates in future experiments. Rational design of adoptive transfer immunotherapy with iTreg cells hinges upon this type of information.
EXPERIMENTAL PROCEDURES
Mice
Foxp3EGFP, Foxp3ΔEGFP, and Foxp3K276X mice on the BALB/c background were generated and screened as described (Haribhai et al., 2007; Lin et al., 2007; Lin et al., 2005). C57BL/6 Rag1−/− mice were obtained from the Jackson Laboratory and backcrossed to BALB/c mice for over 20 generations. Rescued Foxp3K276X/ K276X females and Foxp3K276X males were mated to generate litters where all progeny were Foxp3-deficient. Mice expressing the simian DTR cDNA under the control of foxp3 transcriptional control elements (Foxp3DTR) were generated as described in the supplemental methods. The Animal Resource Committees at the Medical College of Wisconsin and at the University of California, Los Angeles approved all animal experiments.
Cell purification and adoptive transfer
Pooled splenocytes and lymph node cells were stained with either anti-CD4-PE, or anti-CD4-Pacific blue and anti-CD45RB-APC as appropriate, and sorted on the basis of antibody and EGFP fluorescence. All sorting was done on a FACSAria (Becton-Dickenson). The average purity and viability of the sorted CD4+ populations was 98.0±0.2% and 89.5±0.7% respectively (n=49). For the Foxp3-deficient rescue experiments, 200 µl of a cell suspension in PBS was injected into the peritoneal cavity of pups within the first 30 hours following birth.
Diphtheria toxin depletion
Rescued Foxp3ΔEGFP mice (day 50) were given 50µg/kg of diphtheria toxin (Sigma Aldrich) in 200 µl of PBS by intra-peritoneal injection daily or every other day for 15 days. Mice were weighed daily, before each injection when appropriate. Following the last injection, mice were rested for 5 days and then examined. For intracellular cytokine analysis, splenocytes were restimulated in culture with PMA and ionomycin in the 25 presence of Brefeldin A following the manufacture’s instructions (BD Bioscience) for 4 hours, fixed and permeabilized with 0.1% Triton-X, then stained with antibodies as indicated.
Gene expression profiling
The data have been deposited in the NCBI Gene Expression Omnibus repository, accession number GSE19512
Detailed descriptions of the methods for the generation of Foxp3DTR mice, TGF-β1-mediated in vitro conversion, colitis scores, flow cytometry and cytokine analysis, statistics, histology, gene expression analysis, and CDR3 repertoire analysis can be found in the online supplement.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Jane Ebert for technical support, and Richard Lang for providing Simian DTR cDNA. This work was supported by National Institutes of Health grants R01 AI073731 and R01 AI085090 (C.B.W. and T.A.C), N01 50032 (C.B.W.), R01 AI065617 (T.A.C.), R01 AI078713 (M.J.H.), the D.B. and Marjorie Reinhart, and the Montgomery Family Foundations (C.B.W.), the Advancing a Healthier Wisconsin – Research for a Healthier Tomorrow Initiative (C.B.W.), the Crohn’s and Colitis Foundation of America (C.B.W), and the Children’s Hospital of Wisconsin (C.B.W.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Apostolou I, von Boehmer H. In vivo instruction of suppressor commitment in naive T cells. J Exp Med. 2004;199:1401–1408. doi: 10.1084/jem.20040249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunkow ME, Jeffery EW, Hjerrild KA, Paeper B, Clark LB, Yasayko SA, Wilkinson JE, Galas D, Ziegler SF, Ramsdell F. Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nat Genet. 2001;27:68–73. doi: 10.1038/83784. [DOI] [PubMed] [Google Scholar]
- Chatila TA, Blaeser F, Ho N, Lederman HM, Voulgaropoulos C, Helms C, Bowcock AM. JM2, encoding a fork head-related protein, is mutated in X-linked autoimmunity-allergic disregulation syndrome. J Clin Invest. 2000;106:R75–R81. doi: 10.1172/JCI11679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Jin W, Hardegen N, Lei KJ, Li L, Marinos N, McGrady G, Wahl SM. Conversion of peripheral CD4+CD25- naive T cells to CD4+CD25+ regulatory T cells by TGF-beta induction of transcription factor Foxp3. J Exp Med. 2003;198:1875–1886. doi: 10.1084/jem.20030152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coombes JL, Siddiqui KR, Arancibia-Carcamo CV, Hall J, Sun CM, Belkaid Y, Powrie F. A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGF-beta and retinoic acid-dependent mechanism. J Exp Med. 2007;204:1757–1764. doi: 10.1084/jem.20070590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curotto de Lafaille MA, Kutchukhidze N, Shen S, Ding Y, Yee H, Lafaille JJ. Adaptive Foxp3+ regulatory T cell-dependent and -independent control of allergic inflammation. Immunity. 2008;29:114–126. doi: 10.1016/j.immuni.2008.05.010. [DOI] [PubMed] [Google Scholar]
- Curotto de Lafaille MA, Lafaille JJ. Natural and adaptive foxp3+ regulatory T cells: more of the same or a division of labor? Immunity. 2009;30:626–635. doi: 10.1016/j.immuni.2009.05.002. [DOI] [PubMed] [Google Scholar]
- Curotto de Lafaille MA, Lino AC, Kutchukhidze N, Lafaille JJ. CD25- T cells generate CD25+Foxp3+ regulatory T cells by peripheral expansion. J Immunol. 2004;173:7259–7268. doi: 10.4049/jimmunol.173.12.7259. [DOI] [PubMed] [Google Scholar]
- Fantini MC, Becker C, Tubbe I, Nikolaev A, Lehr HA, Galle P, Neurath MF. Transforming growth factor beta induced FoxP3+ regulatory T cells suppress Th1 mediated experimental colitis. Gut. 2006;55:671–680. doi: 10.1136/gut.2005.072801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuerer M, Hill JA, Kretschmer K, von Boehmer H, Mathis D, Benoist C. Genomic definition of multiple ex vivo regulatory T cell subphenotypes. Proc Natl Acad Sci U S A. 2010;107:5919–5924. doi: 10.1073/pnas.1002006107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floess S, Freyer J, Siewert C, Baron U, Olek S, Polansky J, Schlawe K, Chang HD, Bopp T, Schmitt E, et al. Epigenetic control of the foxp3 locus in regulatory T cells. PLoS Biol. 2007;5:e38. doi: 10.1371/journal.pbio.0050038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontenot JD, Gavin MA, Rudensky AY. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat Immunol. 2003;4:330–336. doi: 10.1038/ni904. [DOI] [PubMed] [Google Scholar]
- Fontenot JD, Rasmussen JP, Williams LM, Dooley JL, Farr AG, Rudensky AY. Regulatory T cell lineage specification by the forkhead transcription factor foxp3. Immunity. 2005;22:329–341. doi: 10.1016/j.immuni.2005.01.016. [DOI] [PubMed] [Google Scholar]
- Francisco LM, Salinas VH, Brown KE, Vanguri VK, Freeman GJ, Kuchroo VK, Sharpe AH. PD-L1 regulates the development, maintenance, and function of induced regulatory T cells. J Exp Med. 2009;206:3015–3029. doi: 10.1084/jem.20090847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haribhai D, Lin W, Edwards B, Ziegelbauer J, Salzman NH, Carlson MR, Li SH, Simpson PM, Chatila TA, Williams CB. A central role for induced regulatory T cells in tolerance induction in experimental colitis. J Immunol. 2009;182:3461–3468. doi: 10.4049/jimmunol.0802535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haribhai D, Lin W, Relland LM, Truong N, Williams CB, Chatila TA. Regulatory T cells dynamically control the primary immune response to foreign antigen. J Immunol. 2007;178:2961–2972. doi: 10.4049/jimmunol.178.5.2961. [DOI] [PubMed] [Google Scholar]
- Hill JA, Feuerer M, Tash K, Haxhinasto S, Perez J, Melamed R, Mathis D, Benoist C. Foxp3 transcription-factor-dependent and -independent regulation of the regulatory T cell transcriptional signature. Immunity. 2007;27:786–800. doi: 10.1016/j.immuni.2007.09.010. [DOI] [PubMed] [Google Scholar]
- Hsieh CS, Liang Y, Tyznik AJ, Self SG, Liggitt D, Rudensky AY. Recognition of the peripheral self by naturally arising CD25+ CD4+ T cell receptors. Immunity. 2004;21:267–277. doi: 10.1016/j.immuni.2004.07.009. [DOI] [PubMed] [Google Scholar]
- Hsieh CS, Zheng Y, Liang Y, Fontenot JD, Rudensky AY. An intersection between the self-reactive regulatory and nonregulatory T cell receptor repertoires. Nat Immunol. 2006;7:401–410. doi: 10.1038/ni1318. [DOI] [PubMed] [Google Scholar]
- Huter EN, Punkosdy GA, Glass DD, Cheng LI, Ward JM, Shevach EM. TGF-beta-induced Foxp3+ regulatory T cells rescue scurfy mice. Eur J Immunol. 2008a;38:1814–1821. doi: 10.1002/eji.200838346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huter EN, Stummvoll GH, DiPaolo RJ, Glass DD, Shevach EM. Cutting edge: antigen-specific TGF beta-induced regulatory T cells suppress Th17-mediated autoimmune disease. J Immunol. 2008b;181:8209–8213. doi: 10.4049/jimmunol.181.12.8209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan MS, Boesteanu A, Reed AJ, Petrone AL, Holenbeck AE, Lerman MA, Naji A, Caton AJ. Thymic selection of CD4+CD25+ regulatory T cells induced by an agonist self-peptide. Nat Immunol. 2001;2:301–306. doi: 10.1038/86302. [DOI] [PubMed] [Google Scholar]
- Kedzierska K, Turner SJ, Doherty PC. Conserved T cell receptor usage in primary and recall responses to an immunodominant influenza virus nucleoprotein epitope. Proc Natl Acad Sci U S A. 2004;101:4942–4947. doi: 10.1073/pnas.0401279101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JM, Rasmussen JP, Rudensky AY. Regulatory T cells prevent catastrophic autoimmunity throughout the lifespan of mice. Nat Immunol. 2007;8:191–197. doi: 10.1038/ni1428. [DOI] [PubMed] [Google Scholar]
- Koch MA, Tucker-Heard G, Perdue NR, Killebrew JR, Urdahl KB, Campbell DJ. The transcription factor T-bet controls regulatory T cell homeostasis and function during type 1 inflammation. Nat Immunol. 2009;10:595–602. doi: 10.1038/ni.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lathrop SK, Santacruz NA, Pham D, Luo J, Hsieh CS. Antigen-specific peripheral shaping of the natural regulatory T cell population. J Exp Med. 2008;205:3105–3117. doi: 10.1084/jem.20081359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W, Haribhai D, Relland LM, Truong N, Carlson MR, Williams CB, Chatila TA. Regulatory T cell development in the absence of functional Foxp3. Nat Immunol. 2007;8:359–368. doi: 10.1038/ni1445. [DOI] [PubMed] [Google Scholar]
- Lin W, Truong N, Grossman WJ, Haribhai D, Williams CB, Wang J, Martin MG, Chatila TA. Allergic dysregulation and hyperimmunoglobulinemia E in Foxp3 mutant mice. J Allergy Clin Immunol. 2005;116:1106–1115. doi: 10.1016/j.jaci.2005.08.046. [DOI] [PubMed] [Google Scholar]
- Menezes JS, van den Elzen P, Thornes J, Huffman D, Droin NM, Maverakis E, Sercarz EE. A public T cell clonotype within a heterogeneous autoreactive repertoire is dominant in driving EAE. J Clin Invest. 2007;117:2176–2185. doi: 10.1172/JCI28277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyara M, Amoura Z, Parizot C, Badoual C, Dorgham K, Trad S, Kambouchner M, Valeyre D, Chapelon-Abric C, Debre P, et al. The immune paradox of sarcoidosis and regulatory T cells. J Exp Med. 2006;203:359–370. doi: 10.1084/jem.20050648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottet C, Uhlig HH, Powrie F. Cutting edge: cure of colitis by CD4+CD25+ regulatory T cells. J Immunol. 2003;170:3939–3943. doi: 10.4049/jimmunol.170.8.3939. [DOI] [PubMed] [Google Scholar]
- Mucida D, Kutchukhidze N, Erazo A, Russo M, Lafaille JJ, Curotto de Lafaille MA. Oral tolerance in the absence of naturally occurring Tregs. J Clin Invest. 2005;115:1923–1933. doi: 10.1172/JCI24487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumov YN, Naumova EN, Clute SC, Watkin LB, Kota K, Gorski J, Selin LK. Complex T cell memory repertoires participate in recall responses at extremes of antigenic load. J Immunol. 2006;177:2006–2014. doi: 10.4049/jimmunol.177.3.2006. [DOI] [PubMed] [Google Scholar]
- Pacholczyk R, Ignatowicz H, Kraj P, Ignatowicz L. Origin and T cell receptor diversity of Foxp3+CD4+CD25+ T cells. Immunity. 2006;25:249–259. doi: 10.1016/j.immuni.2006.05.016. [DOI] [PubMed] [Google Scholar]
- Polansky JK, Kretschmer K, Freyer J, Floess S, Garbe A, Baron U, Olek S, Hamann A, von Boehmer H, Huehn J. DNA methylation controls Foxp3 gene expression. Eur J Immunol. 2008;38:1654–1663. doi: 10.1002/eji.200838105. [DOI] [PubMed] [Google Scholar]
- Quigley MF, Greenaway HY, Venturi V, Lindsay R, Quinn KM, Seder RA, Douek DC, Davenport MP, Price DA. Convergent recombination shapes the clonotypic landscape of the naive T-cell repertoire. Proc Natl Acad Sci U S A. 2010;107:19414–19419. doi: 10.1073/pnas.1010586107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana FJ, Basso AS, Iglesias AH, Korn T, Farez MF, Bettelli E, Caccamo M, Oukka M, Weiner HL. Control of T(reg) and T(H)17 cell differentiation by the aryl hydrocarbon receptor. Nature. 2008;453:65–71. doi: 10.1038/nature06880. [DOI] [PubMed] [Google Scholar]
- Relland LM, Mishra MK, Haribhai D, Edwards B, Ziegelbauer J, Williams CB. Affinity-based selection of regulatory T cells occurs independent of agonist-mediated induction of Foxp3 expression. J Immunol. 2009;182:1341–1350. doi: 10.4049/jimmunol.182.3.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schallenberg S, Tsai PY, Riewaldt J, Kretschmer K. Identification of an immediate Foxp3(−) precursor to Foxp3(+) regulatory T cells in peripheral lymphoid organs of nonmanipulated mice. J Exp Med. 2010;207:1393–1407. doi: 10.1084/jem.20100045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz-Winnenthal H, Pietsch DH, Schimmack S, Bonertz A, Udonta F, Ge Y, Galindo L, Volk C, Zgraggen K, Koch M, et al. Chronic pancreatitis is associated with disease-specific regulatory T-cell responses. Gastroenterology. 2009 doi: 10.1053/j.gastro.2009.11.011. [DOI] [PubMed] [Google Scholar]
- Selvaraj RK, Geiger TL. A kinetic and dynamic analysis of Foxp3 induced in T cells by TGF-beta. J Immunol. 2007;178:7667–7677. doi: 10.4049/jimmunol.178.12.7667. [DOI] [PubMed] [Google Scholar]
- Venturi V, Price DA, Douek DC, Davenport MP. The molecular basis for public T-cell responses? Nat Rev Immunol. 2008;8:231–238. doi: 10.1038/nri2260. [DOI] [PubMed] [Google Scholar]
- Wong J, Obst R, Correia-Neves M, Losyev G, Mathis D, Benoist C. Adaptation of TCR repertoires to self-peptides in regulatory and nonregulatory CD4+ T cells. J Immunol. 2007;178:7032–7041. doi: 10.4049/jimmunol.178.11.7032. [DOI] [PubMed] [Google Scholar]
- Yang XO, Nurieva R, Martinez GJ, Kang HS, Chung Y, Pappu BP, Shah B, Chang SH, Schluns KS, Watowich SS, et al. Molecular antagonism and plasticity of regulatory and inflammatory T cell programs. Immunity. 2008;29:44–56. doi: 10.1016/j.immuni.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Bailey-Bucktrout SL, Jeker LT, Penaranda C, Martinez-Llordella M, Ashby M, Nakayama M, Rosenthal W, Bluestone JA. Instability of the transcription factor Foxp3 leads to the generation of pathogenic memory T cells in vivo. Nat Immunol. 2009;10:1000–1007. doi: 10.1038/ni.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.